| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
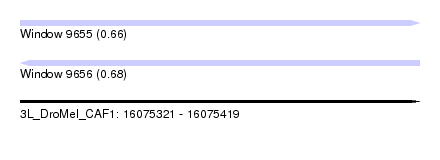
| Location | 16,075,321 – 16,075,419 |
| Length | 98 |
| Max. P | 0.683453 |

| Location | 16,075,321 – 16,075,419 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 79.32 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -21.74 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.659707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
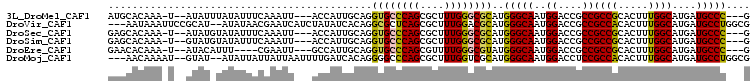
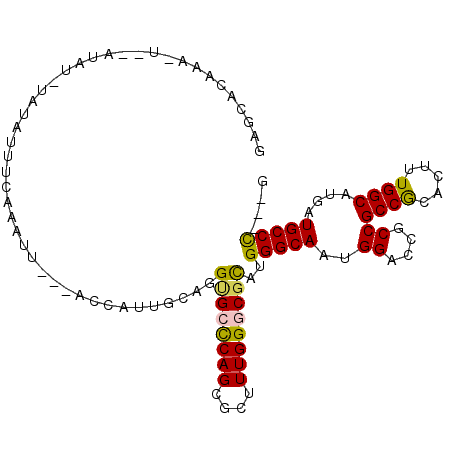
>3L_DroMel_CAF1 16075321 98 + 23771897 C---GGGCAUCAUGCCAAAGUGCGGCGGCGGUCCAUUGCCCAUGCGCCCAAAGCGCUGGGCACCUGCAAUGGU---AAUUUGAAAUAUAAAUAU--A-UUUGUGCAU .---..((((..(((((...(((((.((....))..((((((.((((.....)))))))))).))))).))))---).....((((((....))--)-))))))).. ( -33.70) >DroVir_CAF1 4205 102 + 1 CGCCAGGCAUCAUGCCAAAGUGUGGCGGCGGUCCAUUGCCCAUGCGUCCAAAGCGCUGAGCGCCUGUGAUAUAGAUGAUUCGUUAUAU--AUGCGGAAUUUAUU--- (((.((((.(((.((((.....))))(((((....)))))...((((.....)))))))..)))))))..((((((..(((((.....--..))))))))))).--- ( -28.10) >DroSec_CAF1 4237 98 + 1 C---GGGCAUCAUGCCAAAGUGCGGCGGCGGUCCAUUGCCCAUGCGCCCAAAGCGCUGGGCACCUGCAAUGGU---AAUUUGAAAUAUACAUAU--A-UUUGUGCUC .---((((((..(((((...(((((.((....))..((((((.((((.....)))))))))).))))).))))---).....((((((....))--)-))))))))) ( -35.90) >DroSim_CAF1 10396 98 + 1 C---GGGCAUCAUGCCAAAGUGCGGCGGCGGUCCAUUGCCCAUGCGCCCAAAGCGCUGGGCACCUGCAAUGGU---AAUUUGAAAUAUACAUAC--A-UUUGUGCUC .---((((.((((((((...(((((.((....))..((((((.((((.....)))))))))).))))).))))---)...))).....(((...--.-..))))))) ( -34.20) >DroEre_CAF1 4188 94 + 1 C---GGGCAUCAUGCCAAAGUGCGGCGGCGGUCCAUUGCCCAUACGCCCAAAACGCUGGGCACCUGCAAUGGC---AAUUCG----AAAUGUAU--A-UUUGUGUUC .---((((((..(((((...(((((.(((((....))))).....(((((......)))))..))))).))))---).....----((((....--)-))))))))) ( -28.80) >DroMoj_CAF1 3874 100 + 1 CGCCAGGCAUCAUGCCAAAGUGUGGCGGAGGUCCAUUGCCCAUGCGACCAAAGCGCUGGGCCCCUGUGAUCAAAAUUAAUAAUAAUAU--AUAC--AUUUUGUU--- ((((((((.....)))......)))))..((((((..(((((.(((.......))))))))...)).)))).................--....--........--- ( -27.90) >consensus C___GGGCAUCAUGCCAAAGUGCGGCGGCGGUCCAUUGCCCAUGCGCCCAAAGCGCUGGGCACCUGCAAUGGU___AAUUUGAAAUAUA_AUAC__A_UUUGUGCUC ....((((....((((.......))))...))))..((((((.((((.....))))))))))............................................. (-21.74 = -22.72 + 0.97)

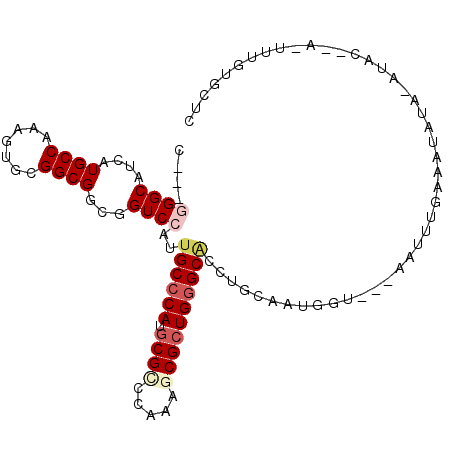
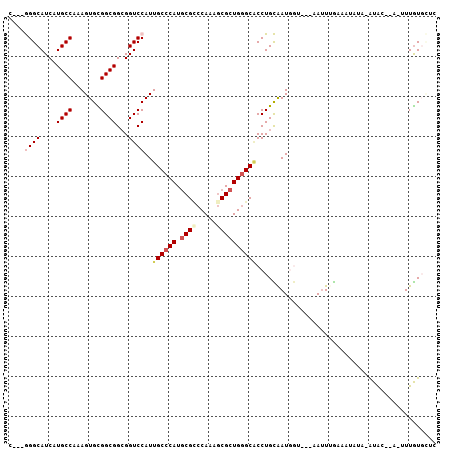
| Location | 16,075,321 – 16,075,419 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.32 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -23.15 |
| Energy contribution | -22.82 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16075321 98 - 23771897 AUGCACAAA-U--AUAUUUAUAUUUCAAAUU---ACCAUUGCAGGUGCCCAGCGCUUUGGGCGCAUGGGCAAUGGACCGCCGCCGCACUUUGGCAUGAUGCCC---G ..(((((..-.--..................---.(((((((..((((((((....))))))))....)))))))......((((.....)))).)).)))..---. ( -32.40) >DroVir_CAF1 4205 102 - 1 ---AAUAAAUUCCGCAU--AUAUAACGAAUCAUCUAUAUCACAGGCGCUCAGCGCUUUGGACGCAUGGGCAAUGGACCGCCGCCACACUUUGGCAUGAUGCCUGGCG ---.........(((..--.......((....)).......((((((((((((((....).))).)))))...((....))((((.....)))).....)))))))) ( -28.30) >DroSec_CAF1 4237 98 - 1 GAGCACAAA-U--AUAUGUAUAUUUCAAAUU---ACCAUUGCAGGUGCCCAGCGCUUUGGGCGCAUGGGCAAUGGACCGCCGCCGCACUUUGGCAUGAUGCCC---G (.(((((..-.--..................---.(((((((..((((((((....))))))))....)))))))......((((.....)))).)).))).)---. ( -33.00) >DroSim_CAF1 10396 98 - 1 GAGCACAAA-U--GUAUGUAUAUUUCAAAUU---ACCAUUGCAGGUGCCCAGCGCUUUGGGCGCAUGGGCAAUGGACCGCCGCCGCACUUUGGCAUGAUGCCC---G (.(((((..-.--..................---.(((((((..((((((((....))))))))....)))))))......((((.....)))).)).))).)---. ( -33.00) >DroEre_CAF1 4188 94 - 1 GAACACAAA-U--AUACAUUU----CGAAUU---GCCAUUGCAGGUGCCCAGCGUUUUGGGCGUAUGGGCAAUGGACCGCCGCCGCACUUUGGCAUGAUGCCC---G .........-.--((((....----......---(((......)))((((((....))))))))))(((((..((....))((((.....))))....)))))---. ( -28.90) >DroMoj_CAF1 3874 100 - 1 ---AACAAAAU--GUAU--AUAUUAUUAUUAAUUUUGAUCACAGGGGCCCAGCGCUUUGGUCGCAUGGGCAAUGGACCUCCGCCACACUUUGGCAUGAUGCCUGGCG ---..((((((--..((--(......)))..))))))....((((.(((((((((....).))).)))))...((....))((((.....))))......))))... ( -29.30) >consensus GAGCACAAA_U__AUAU_UAUAUUUCAAAUU___ACCAUUGCAGGUGCCCAGCGCUUUGGGCGCAUGGGCAAUGGACCGCCGCCGCACUUUGGCAUGAUGCCC___G ............................................((((((((....))))))))..(((((..((....))((((.....))))....))))).... (-23.15 = -22.82 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:25 2006