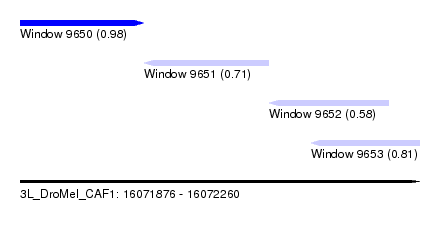
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,071,876 – 16,072,260 |
| Length | 384 |
| Max. P | 0.984164 |

| Location | 16,071,876 – 16,071,995 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.98 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -25.16 |
| Energy contribution | -25.72 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16071876 119 + 23771897 AAAUCCGCUUCGUUCUAGUAUUUGUGUAUAUGAGUGUGAGGGCCAUAAUGUUCACAUUCAAACUGAA-UACAUUGCCUUGCCUUGCACUAAUGCUAGAAAAAUACAAGUUUUAUGAUCCA ......((((..((((((((((.(((((..((((((((((.(......).)))))))))).......-..((......))...))))).))))))))))......))))........... ( -27.80) >DroSec_CAF1 777 118 + 1 AAAUCCGCUUCGUUCUUGUAUUUGUGUAUAUGAGUGUGAGGG-CAUAAUGUUCACAUUCAAACUGAA-UACAUUGCCUUGCCUUGCACUAAUGCUAGAAAAAUACAAGUUUUAUGAUCCA ......(..((((.(((((((((..((((...(((((.((((-((((((((....((((.....)))-)))))))...))))))))))).)))).....)))))))))....))))..). ( -31.00) >DroSim_CAF1 6930 119 + 1 AAAUCCGCUUCGUUCUUGUAUUUGUGUAUAUGAGUGUGAGGGCCAUAAUGUUCACAUUCAAACUGAA-UACAUUGCCUUGCCUUGCACUAAUGCUAGAAAAAUACAAGUUUUAUGAUCCA ......(..((((.(((((((((..((((...((((..(((....((((((....((((.....)))-))))))).....)))..)))).)))).....)))))))))....))))..). ( -29.70) >DroEre_CAF1 806 119 + 1 AAAUCCGCUUUGUUCUUGUAUUUGUGUAAAUGAGUGUGAGGGCCUAAAAGUUCACAUUCAAACUGAA-UACAUUGCCUUGCCUUGCACUAAUGCUAGAAAAAUACAAGUUUUAUGAUCCG .....((..(..(.(((((((((..(((....((((..(((..............((((.....)))-).((......)))))..))))..))).....)))))))))....)..)..)) ( -23.30) >DroYak_CAF1 7039 120 + 1 AAAUCCGCUUUGUUCUUGUAUUUGUGUAAAUGAGUGUGAGGGCCAAAAUGUUCACAUUCAAACUGAAUUACAUUGCCUUGCCUUGCACUAAUGCUAAAAAAAUACAAGUUUUAUGAUCCG .....((..(..(.(((((((((..(((....((((..(((.....(((((....((((.....)))).)))))......)))..))))..))).....)))))))))....)..)..)) ( -26.80) >consensus AAAUCCGCUUCGUUCUUGUAUUUGUGUAUAUGAGUGUGAGGGCCAUAAUGUUCACAUUCAAACUGAA_UACAUUGCCUUGCCUUGCACUAAUGCUAGAAAAAUACAAGUUUUAUGAUCCA ......(..((((.(((((((((..((((...((((..(((.....(((((.....(((.....)))..)))))......)))..)))).)))).....)))))))))....))))..). (-25.16 = -25.72 + 0.56)


| Location | 16,071,995 – 16,072,115 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.34 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16071995 120 - 23771897 CUAAUCUAUACACGAGUAUUCAAACCGAUUGGGGCACAAGGAGCUAAGCAUGUCCAACAUGUUGCCCAAUCGGCGAUUAGUGAAUCCCAAAUUACCCCAACUUUAACUAACAAUAUUUAG ((((((.((((....)))).....(((((((((((.......))..((((((.....)))))).))))))))).))))))........................................ ( -27.60) >DroSec_CAF1 895 120 - 1 CUAAUCUGUACACGAGUAUUCAAACCGAUUGGGGCACAAGUAGCUAAGCAUGUCCAACAUGUUGCCCAAUCGGCGUUUAGUGAAUCCCAAACUACCCCAACUUUAACUAACAAUAUUUAG .............(((((((....(((((((((((.......))..((((((.....)))))).))))))))).(..((((.........))))..)..............))))))).. ( -25.50) >DroSim_CAF1 7049 120 - 1 CUAAUCUGUACACGAGUAUUCAAACCGAUUGGGGCACAAGGAGCUAAGCAUGUCCAACAUGUUGCCCAAUCGGCGUUUAGUGAAUCCCAAACUACCACAACUUUAACUAACAAUAUUUAG .............(((((((....(((((((((((.......))..((((((.....)))))).))))))))).((.((((.........))))..)).............))))))).. ( -25.50) >DroEre_CAF1 925 119 - 1 CUUAUCUGUACACGAGUAUUCAAACCAAUUUGGGCACAAGGAGCUAAGCAUGUCCAACAUGUUGCCCAAUUGGCGAUUAGUGAAUCCCCA-UUACACGAACUUUAACUAACAAUAUUUAG .............(((((((....((((((.(((((...........(((((.....))))))))))))))))((..(((((......))-)))..)).............))))))).. ( -22.90) >DroYak_CAF1 7159 119 - 1 CUUAUCUGUACACGAGUAUUCAAACCAAUUUGGGCACAAGGAGCUAAGCAUGUCCAACAUGUUGCCCAAUUGGCGAUUAGUGAAUCCCCA-AUACACGAACAUUAACUAACCAUAUUUAG ............((.(((((....((((((.(((((...........(((((.....)))))))))))))))).((((....))))...)-)))).))...................... ( -22.60) >consensus CUAAUCUGUACACGAGUAUUCAAACCGAUUGGGGCACAAGGAGCUAAGCAUGUCCAACAUGUUGCCCAAUCGGCGAUUAGUGAAUCCCAAACUACCCCAACUUUAACUAACAAUAUUUAG ((((((.((((....)))).....((((((.(((((...........(((((.....)))))))))))))))).))))))........................................ (-22.18 = -22.34 + 0.16)

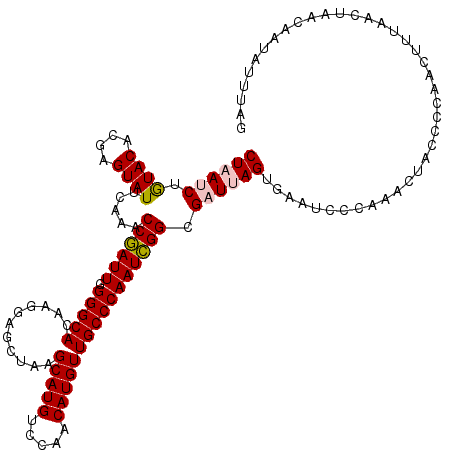
| Location | 16,072,115 – 16,072,230 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -24.94 |
| Consensus MFE | -21.16 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16072115 115 - 23771897 ACUCCUGCCAGCACCUAAGCGGCGAGAAUAGGUUAUAUACACGUAGUCGGCUACUAAUAUUACAAUUUCUCUCAAGAUAAAAGCGCGACCAGGCGUCACUUAACUAUAACCAACU .(((..(((.((......))))))))....(((((((.....((((....))))............................((((......))))........))))))).... ( -23.80) >DroSec_CAF1 1015 115 - 1 ACUCCUGCCAGCACCUAAGCGGCGAGAAUAGGUUAUAUACACGUAGUCGGCUACUAAUAUUACAAUUUCUCUCAAGAUAAAUGCGCGACCAGGCGUCACUUAACUAUAACCAACU .(((..(((.((......))))))))....(((((((.....((((....)))).......................(((.(((((......))).)).)))..))))))).... ( -24.20) >DroSim_CAF1 7169 115 - 1 ACUCCUGCCAGCACCUAAGCGGCGAGAAUAGGUUAUAUACACGUAGUCGGCUACUAAUAUUACAAUUUCUCUCAAGAUAAUUGCGCGAACAGGCGUCACUUAACUAUAACCAACU .(((..(((.((......))))))))....(((((((.....((((....)))).......................(((.(((((......))).)).)))..))))))).... ( -24.20) >DroEre_CAF1 1044 115 - 1 ACUGCUGCCAGUGCAUAGGCGGCGAGAAUAGGUUAUUUACACGUAGUCGGCUACUAAAAUUACAAUUUCUCUCAAGAUAAAUGCGCAUCCAAGCGUCACUUAACUAUAACCAACU ...((((((........))))))(((((...((.((((....((((....))))..)))).))...)))))...((.(((.(((((......))).)).))).)).......... ( -27.00) >DroYak_CAF1 7278 115 - 1 ACUCCUGCCAGUGCCUAAGCGGCGGGAAUAGAUUAUAUACACGUAGUCGGCUACUAAAAUUACAAUUUCUCUUAAGAUAAAUGCGCGUCCAAGCGCCACUUAACUAUAACCAACU ..(((((((.((......)))))))))((((...........((((....)))).................(((((......((((......))))..)))))))))........ ( -25.50) >consensus ACUCCUGCCAGCACCUAAGCGGCGAGAAUAGGUUAUAUACACGUAGUCGGCUACUAAUAUUACAAUUUCUCUCAAGAUAAAUGCGCGACCAGGCGUCACUUAACUAUAACCAACU .(((..(((.((......))))))))....(((((((.....((((....))))............................((((......))))........))))))).... (-21.16 = -21.20 + 0.04)

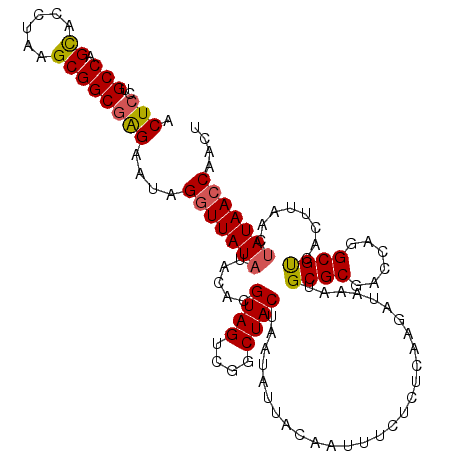

| Location | 16,072,155 – 16,072,260 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 94.10 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.16 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16072155 105 - 23771897 AAGGUGGCCAAGAAGGAGGGUGUCGGAGGCACUCCUGCCAGCACCUAAGCGGCGAGAAUAGGUUAUAUACACGUAGUCGGCUACUAAUAUUACAAUUUCUCUCAA ......(((.....(((((((((.....)))).))).)).((......)))))(((((...((.((((....((((....))))..)))).))...))))).... ( -29.60) >DroSec_CAF1 1055 105 - 1 AAGGUGGCCAAGAAGGAGGGUGCCGGAGUCACUCCUGCCAGCACCUAAGCGGCGAGAAUAGGUUAUAUACACGUAGUCGGCUACUAAUAUUACAAUUUCUCUCAA ..((((.((........)).))))((((...)))).(((.((......)))))(((((...((.((((....((((....))))..)))).))...))))).... ( -28.50) >DroSim_CAF1 7209 105 - 1 AAGGUGGCCAAGAAGGAGGGUGCUGGAGGCACUCCUGCCAGCACCUAAGCGGCGAGAAUAGGUUAUAUACACGUAGUCGGCUACUAAUAUUACAAUUUCUCUCAA ..(((((((........((((((((((((....))).)))))))))..((.(((.(.(((.....))).).))).)).))))))).................... ( -34.60) >DroEre_CAF1 1084 105 - 1 AAGGUGGCCAAGAAGGAGGGUGCCGGAGGCACUGCUGCCAGUGCAUAGGCGGCGAGAAUAGGUUAUUUACACGUAGUCGGCUACUAAAAUUACAAUUUCUCUCAA ..(((((((.........((((((...))))))((((((........)))))).......(..(((......)))..)))))))).................... ( -32.20) >DroYak_CAF1 7318 105 - 1 AAAGUGGCCAAGAAGGAGGGUGCCGGAGGCACUCCUGCCAGUGCCUAAGCGGCGGGAAUAGAUUAUAUACACGUAGUCGGCUACUAAAAUUACAAUUUCUCUUAA ..(((((((..........(((((...)))))(((((((.((......)))))))))...((((((......))))))))))))).................... ( -34.80) >consensus AAGGUGGCCAAGAAGGAGGGUGCCGGAGGCACUCCUGCCAGCACCUAAGCGGCGAGAAUAGGUUAUAUACACGUAGUCGGCUACUAAUAUUACAAUUUCUCUCAA ..(((((((.....((.(((((((...)))))))))(((.((......))))).......((((((......))))))))))))).................... (-26.60 = -26.16 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:22 2006