| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,071,103 – 16,071,214 |
| Length | 111 |
| Max. P | 0.991131 |

| Location | 16,071,103 – 16,071,214 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.96 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
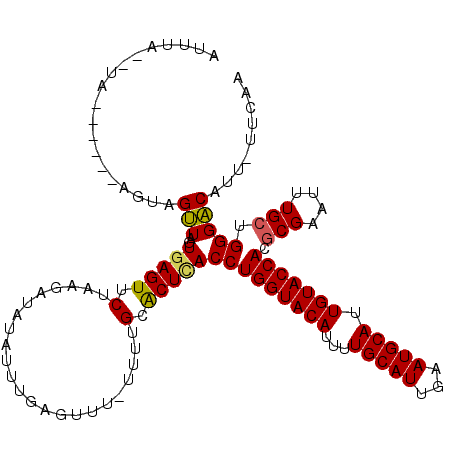
>3L_DroMel_CAF1 16071103 111 + 23771897 UUGAAUAAUGUCCCAGCAAAUUCGCGUGGUACAAUGCAUUCAAUGCAAAAUGUACCAGGUGAGUGCAAAC-AAACUGAAAUAUAUCUUAGAACUCAAUAACUACU------UA--UAUAU ((((...........(((...((((.(((((((.(((((...)))))...))))))).)))).)))....-...((((........))))...))))........------..--..... ( -25.50) >DroSec_CAF1 1 111 + 1 -------AUGUCCCAGCAAAUUCGCGUGGUACAAUGCAUUCAAUGCAAAAUGUACCAGGUGAGUGCAAAAAAAAUUUAAAUAUUUCUUAGAACUCACUAACUACUACUACUAC--UAAAU -------........(((...((((.(((((((.(((((...)))))...))))))).)))).)))....................((((......)))).............--..... ( -23.40) >DroSim_CAF1 6156 117 + 1 UUGAAAAAUGUCCCCGCAAAUUCGUGUGGUACAAUGCAUUCAAUGCAAAAUGUACCAGGUGAGUGCAAAA-AAACUAAAAUAUAUCCUAGAACUCACUAACUACUACUACUAC--UAAAU ..............(((......)))(((((((.(((((...)))))...)))))))(((((((......-....................)))))))...............--..... ( -20.47) >DroEre_CAF1 1 104 + 1 -------AUGUCCCAGCAAAUUCGCGUGGUACAAUGCAUUCAAUGCAAAAUGUACCAGGUUAGCGCAUAA--A-CUCCAUUAAACAUCAGAACUCAUCAACUACU------UACAUACAU -------........((......((.(((((((.(((((...)))))...))))))).))....))....--.-...............................------......... ( -16.20) >DroYak_CAF1 6234 106 + 1 UUGAAAAAUGCCCCAGCAAAUUCGCGUGGUACAAUGCAUCCAAUGCAAAAUGUACCAGGUAAGCGCACAA--AUCUCAAGUAAUCCUUAGAACUCAAUAGCUACU------UAA------ ..(((...(((....)))..)))((.(((((((.(((((...)))))...))))))).)).(((......--.(((.(((.....))))))........)))...------...------ ( -21.16) >consensus UUGAA_AAUGUCCCAGCAAAUUCGCGUGGUACAAUGCAUUCAAUGCAAAAUGUACCAGGUGAGUGCAAAA_AAACUCAAAUAUAUCUUAGAACUCAAUAACUACU______UA__UAAAU ...............((..((((((.(((((((.(((((...)))))...))))))).))))))))...................................................... (-19.32 = -19.96 + 0.64)


| Location | 16,071,103 – 16,071,214 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -18.10 |
| Energy contribution | -17.82 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16071103 111 - 23771897 AUAUA--UA------AGUAGUUAUUGAGUUCUAAGAUAUAUUUCAGUUU-GUUUGCACUCACCUGGUACAUUUUGCAUUGAAUGCAUUGUACCACGCGAAUUUGCUGGGACAUUAUUCAA .((((--(.------..(((..........)))..)))))(..((((..-((((((.......(((((((...(((((...))))).))))))).))))))..))))..).......... ( -26.90) >DroSec_CAF1 1 111 - 1 AUUUA--GUAGUAGUAGUAGUUAGUGAGUUCUAAGAAAUAUUUAAAUUUUUUUUGCACUCACCUGGUACAUUUUGCAUUGAAUGCAUUGUACCACGCGAAUUUGCUGGGACAU------- .....--((..((((((..((..((((((.(.(((((((......)))))))..).)))))).(((((((...(((((...))))).))))))).))....))))))..))..------- ( -29.00) >DroSim_CAF1 6156 117 - 1 AUUUA--GUAGUAGUAGUAGUUAGUGAGUUCUAGGAUAUAUUUUAGUUU-UUUUGCACUCACCUGGUACAUUUUGCAUUGAAUGCAUUGUACCACACGAAUUUGCGGGGACAUUUUUCAA .....--((((.....((.....((((((.(.(((((........))))-)...).)))))).(((((((...(((((...))))).))))))).))....))))(....)......... ( -24.80) >DroEre_CAF1 1 104 - 1 AUGUAUGUA------AGUAGUUGAUGAGUUCUGAUGUUUAAUGGAG-U--UUAUGCGCUAACCUGGUACAUUUUGCAUUGAAUGCAUUGUACCACGCGAAUUUGCUGGGACAU------- ((((..(((------(((.....((((..(((..........))).-.--))))(((......(((((((...(((((...))))).))))))))))..))))))....))))------- ( -23.70) >DroYak_CAF1 6234 106 - 1 ------UUA------AGUAGCUAUUGAGUUCUAAGGAUUACUUGAGAU--UUGUGCGCUUACCUGGUACAUUUUGCAUUGGAUGCAUUGUACCACGCGAAUUUGCUGGGGCAUUUUUCAA ------.((------(((.((...(((((..((((.....))))..))--))).)))))))..(((((((...(((((...))))).)))))))...(((..(((....)))...))).. ( -27.80) >consensus AUUUA__UA______AGUAGUUAGUGAGUUCUAAGAUAUAUUUGAGUUU_UUUUGCACUCACCUGGUACAUUUUGCAUUGAAUGCAUUGUACCACGCGAAUUUGCUGGGACAUU_UUCAA ...................(((..(((((.(.......................).)))))(((((((((...(((((...))))).))))))).(((....))).)))))......... (-18.10 = -17.82 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:17 2006