
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,066,818 – 16,067,018 |
| Length | 200 |
| Max. P | 0.929987 |

| Location | 16,066,818 – 16,066,938 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -28.42 |
| Energy contribution | -28.87 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16066818 120 - 23771897 UUCCCUGAUCCAACACUGAUUUUCCGCGUAGAAAAUCAGGAGCUUCCUUGGUGUAAGUGCCCUCCUCAUUCACAAAACACUUUACAGGAAUUUUCUGGGUCAGACUAACAAGGAAAUCUU ((((((((((((...(((((((((......)))))))))(((.(((((.(((((..(((...........)))...)))))....)))))..)))))))))))........))))..... ( -32.10) >DroSim_CAF1 1884 120 - 1 UUCCCUGAUCCAACACUGAUUUUCCGCGUAGAAAAUCAGGAGCUUCCGUGGUGUAAGUGCCCUCCUCAUUCACAAAACAUUUUACAGGAAUUUUCUGGGUCAGACUAACAAGGAAAUCUU ((((((((((((...(((((((((......)))))))))(((.((((...(((.(((((..................)))))))).))))..)))))))))))........))))..... ( -29.37) >DroYak_CAF1 1946 120 - 1 UUCCCUGAUCCAACACUGAUUGUCCGCGUAGAAAAUCAGGAGCUUCCUUGGUGUAAGUGCCCUCCUCAUUCACAAAACAUUUAACAGGAAUUUUUUGGGUCAGACUAACAAGGAAAUCUU (((((((((((((..((((((.((......)).))))))....(((((.(((((..(((...........)))...)))))....)))))....)))))))))........))))..... ( -26.60) >consensus UUCCCUGAUCCAACACUGAUUUUCCGCGUAGAAAAUCAGGAGCUUCCUUGGUGUAAGUGCCCUCCUCAUUCACAAAACAUUUUACAGGAAUUUUCUGGGUCAGACUAACAAGGAAAUCUU ((((((((((((...(((((((((......)))))))))....(((((.(((((..(((...........)))...)))))....))))).....))))))))........))))..... (-28.42 = -28.87 + 0.45)



| Location | 16,066,858 – 16,066,978 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -37.27 |
| Energy contribution | -36.83 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.36 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16066858 120 + 23771897 GUGUUUUGUGAAUGAGGAGGGCACUUACACCAAGGAAGCUCCUGAUUUUCUACGCGGAAAAUCAGUGUUGGAUCAGGGAAAUUCCCUAGUCCUCGACCGCAUAGCAGAGGAUGUAGUCCA .((((.((((..((((((.((((((.......(((.....)))((((((((....)))))))))))))).....((((.....))))..))))))..)))).))))..((((...)))). ( -39.20) >DroSim_CAF1 1924 120 + 1 AUGUUUUGUGAAUGAGGAGGGCACUUACACCACGGAAGCUCCUGAUUUUCUACGCGGAAAAUCAGUGUUGGAUCAGGGAAAUUCCCUAAUCCUCGACCGCAUAGCAGAGGAUGUAGUCCA .((((.((((..((((((((.........))..((((..(((((((((...((((.(.....).)))).)))))))))...))))....))))))..)))).))))..((((...)))). ( -39.00) >DroYak_CAF1 1986 120 + 1 AUGUUUUGUGAAUGAGGAGGGCACUUACACCAAGGAAGCUCCUGAUUUUCUACGCGGACAAUCAGUGUUGGAUCAGGGAAAUUCCCUAGUCCUUGAACGCAUAGCAGAGGAUGUGGUUCA .((((.((((..((((((((.........))..((((..(((((((((...((((.(.....).)))).)))))))))...))))....))))))..)))).)))).............. ( -32.90) >consensus AUGUUUUGUGAAUGAGGAGGGCACUUACACCAAGGAAGCUCCUGAUUUUCUACGCGGAAAAUCAGUGUUGGAUCAGGGAAAUUCCCUAGUCCUCGACCGCAUAGCAGAGGAUGUAGUCCA .((((.((((..((((((((.........))..((((..(((((((((...((((.(.....).)))).)))))))))...))))....))))))..)))).))))..((((...)))). (-37.27 = -36.83 + -0.44)

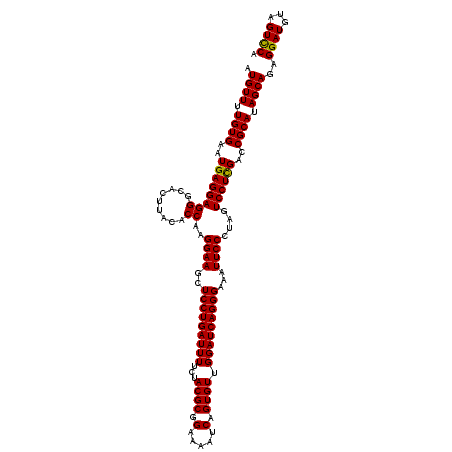
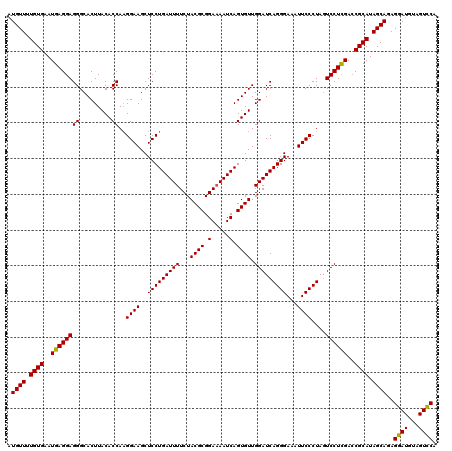
| Location | 16,066,898 – 16,067,018 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -35.55 |
| Energy contribution | -35.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16066898 120 + 23771897 CCUGAUUUUCUACGCGGAAAAUCAGUGUUGGAUCAGGGAAAUUCCCUAGUCCUCGACCGCAUAGCAGAGGAUGUAGUCCACUCCUCAAAGCUGGAACAUUCCUAUCCUAUUGAUUGGCGC .((((((((((....))))))))))....((((.((((.....)))).)))).....(((....(((((((((...((((((......)).)))).......)))))).)))....))). ( -37.10) >DroSim_CAF1 1964 120 + 1 CCUGAUUUUCUACGCGGAAAAUCAGUGUUGGAUCAGGGAAAUUCCCUAAUCCUCGACCGCAUAGCAGAGGAUGUAGUCCACUCCUCCAAGCUGGAACAUUCCUAUCCCAUUGAUUGGCGC .((((((((((....))))))))))....((((.((((.....)))).)))).....(((..(((.(((((.((.....)))))))...)))(((....)))..............))). ( -35.80) >DroYak_CAF1 2026 120 + 1 CCUGAUUUUCUACGCGGACAAUCAGUGUUGGAUCAGGGAAAUUCCCUAGUCCUUGAACGCAUAGCAGAGGAUGUGGUUCACUCCUCCAAGCUGGAGCAUUCCUAUCCCAUAGAUUGGCGU ...........((((...(((((.(((((((((.((((.....)))).))))...)))))......(.(((((.((....((((........))))....))))))))...))))))))) ( -37.50) >consensus CCUGAUUUUCUACGCGGAAAAUCAGUGUUGGAUCAGGGAAAUUCCCUAGUCCUCGACCGCAUAGCAGAGGAUGUAGUCCACUCCUCCAAGCUGGAACAUUCCUAUCCCAUUGAUUGGCGC .((((((((((....))))))))))....((((.((((.....)))).)))).....(((..(((.(((((.((.....)))))))...)))(((....)))..............))). (-35.55 = -35.67 + 0.11)

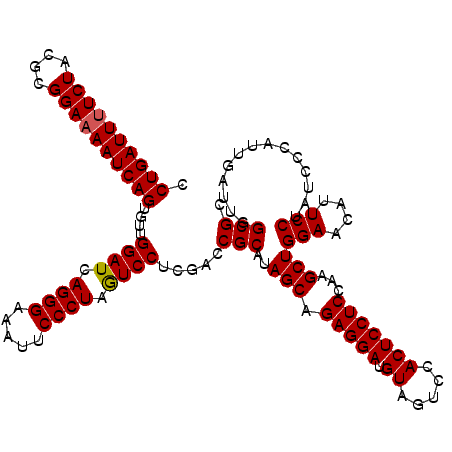
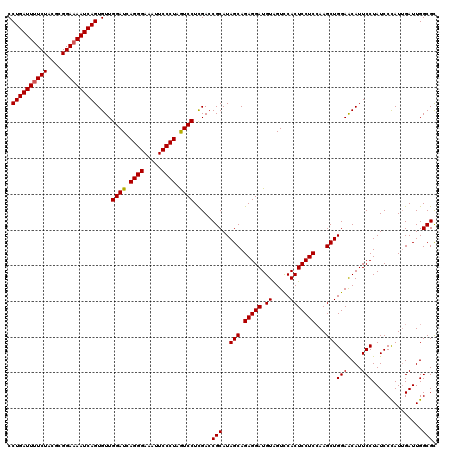
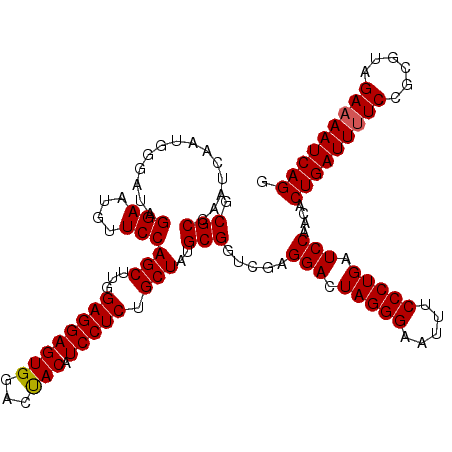
| Location | 16,066,898 – 16,067,018 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -44.27 |
| Consensus MFE | -37.59 |
| Energy contribution | -37.70 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16066898 120 - 23771897 GCGCCAAUCAAUAGGAUAGGAAUGUUCCAGCUUUGAGGAGUGGACUACAUCCUCUGCUAUGCGGUCGAGGACUAGGGAAUUUCCCUGAUCCAACACUGAUUUUCCGCGUAGAAAAUCAGG .((((.............(((....)))(((...(((((.((.....))))))).)))....)).)).(((.(((((.....))))).)))....(((((((((......))))))))). ( -39.70) >DroSim_CAF1 1964 120 - 1 GCGCCAAUCAAUGGGAUAGGAAUGUUCCAGCUUGGAGGAGUGGACUACAUCCUCUGCUAUGCGGUCGAGGAUUAGGGAAUUUCCCUGAUCCAACACUGAUUUUCCGCGUAGAAAAUCAGG .((((.......((((((....))))))(((..((((((.((.....)))))))))))....)).)).(((((((((.....)))))))))....(((((((((......))))))))). ( -47.30) >DroYak_CAF1 2026 120 - 1 ACGCCAAUCUAUGGGAUAGGAAUGCUCCAGCUUGGAGGAGUGAACCACAUCCUCUGCUAUGCGUUCAAGGACUAGGGAAUUUCCCUGAUCCAACACUGAUUGUCCGCGUAGAAAAUCAGG .......((((((((((((((((((...(((..(((((((((...))).)))))))))..))))))..(((.(((((.....))))).)))........)))))).))))))........ ( -45.80) >consensus GCGCCAAUCAAUGGGAUAGGAAUGUUCCAGCUUGGAGGAGUGGACUACAUCCUCUGCUAUGCGGUCGAGGACUAGGGAAUUUCCCUGAUCCAACACUGAUUUUCCGCGUAGAAAAUCAGG .(((..............(((....)))(((...((((((((...))).))))).)))..))).....(((.(((((.....))))).)))....(((((((((......))))))))). (-37.59 = -37.70 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:11 2006