| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
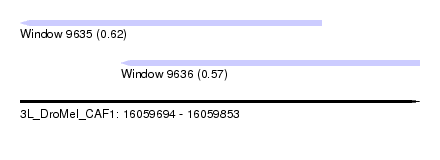
| Location | 16,059,694 – 16,059,853 |
| Length | 159 |
| Max. P | 0.620044 |

| Location | 16,059,694 – 16,059,814 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -35.89 |
| Energy contribution | -35.23 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.37 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
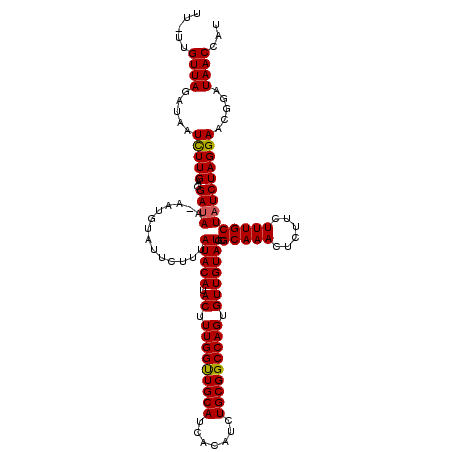
>3L_DroMel_CAF1 16059694 120 - 23771897 UACAUACUUUGGUUGCAUCACAUCUGCGGCCAGUGUUGUAUGCGCAAACUCUUCUUUGCUGUCUAGGAACGGAUAACCAUUCAGCGCAAUGUGCGCAUCGUAGAUACCUUGCCUGCCCAG ((((.((.(((((((((.......))))))))).)))))).(((((((......)))))((((((.((..((....)).....((((.....)))).)).))))))....))........ ( -34.90) >DroSim_CAF1 65 120 - 1 UACAUACUUUGGUUGCAUCACAUCUGCGGCCAGUGUUGUAUGCGCAAACUCUUCUUUGCUAUCUAGGAACGGAUAACCAUUCAGCGCUAUGUGCGCAUCGUAGAUACCUUGCCUGCCGAG ((((.((.(((((((((.......))))))))).)))))).(((((((......)))))((((((.((..((....)).....((((.....)))).)).))))))....))........ ( -35.10) >DroYak_CAF1 241 120 - 1 UACAUACUUUGGCUGCAUCACAUCUGCGGCCAGUGUUGUAUGCGCAAACUCUUCUUUGCUAUCUAGGAACGGAUAACCAUUCAGCGCUAUGUGUGCAUCGUAGAUACCUUGCCUGCCGAG ((((.((.(((((((((.......))))))))).)))))).(((((((......)))))((((((.((..((....)).....((((.....)))).)).))))))....))........ ( -35.70) >consensus UACAUACUUUGGUUGCAUCACAUCUGCGGCCAGUGUUGUAUGCGCAAACUCUUCUUUGCUAUCUAGGAACGGAUAACCAUUCAGCGCUAUGUGCGCAUCGUAGAUACCUUGCCUGCCGAG ((((.((.(((((((((.......))))))))).)))))).(((((((......)))))((((((.((..((....)).....((((.....)))).)).))))))....))........ (-35.89 = -35.23 + -0.66)



| Location | 16,059,734 – 16,059,853 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.15 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.23 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16059734 119 - 23771897 UU-UUGUUAGAUGAUCUUGAGGAAAAAAAGGUUUUCUUUAUACAUACUUUGGUUGCAUCACAUCUGCGGCCAGUGUUGUAUGCGCAAACUCUUCUUUGCUGUCUAGGAACGGAUAACCAU ..-((.((((((.....(((((((((.....)))))))))..(((((.(((((((((.......)))))))))....))))).(((((......))))).)))))).)).((....)).. ( -29.50) >DroSim_CAF1 105 119 - 1 UAGAAGUUAGUUUAUCUUGAGGAUAA-AAUGUGUUAUUAAUACAUACUUUGGUUGCAUCACAUCUGCGGCCAGUGUUGUAUGCGCAAACUCUUCUUUGCUAUCUAGGAACGGAUAACCAU .(((((....((((((.....)))))-).(((((.....(((((.((.(((((((((.......))))))))).))))))))))))....)))))....(((((......)))))..... ( -28.20) >DroYak_CAF1 281 118 - 1 AU-UUGUUAGAUAAUUUUGAUGAUAA-UUUUUAUACCUUAUACAUACUUUGGCUGCAUCACAUCUGCGGCCAGUGUUGUAUGCGCAAACUCUUCUUUGCUAUCUAGGAACGGAUAACCAU ..-((.(((((((.............-...........((((((.((.(((((((((.......))))))))).)))))))).(((((......)))))))))))).)).((....)).. ( -29.40) >consensus UU_UUGUUAGAUAAUCUUGAGGAUAA_AAUGUAUUCUUUAUACAUACUUUGGUUGCAUCACAUCUGCGGCCAGUGUUGUAUGCGCAAACUCUUCUUUGCUAUCUAGGAACGGAUAACCAU .....((((.....(((((..((((..............(((((.((.(((((((((.......))))))))).)))))))..(((((......)))))))))))))).....))))... (-24.34 = -24.23 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:05 2006