
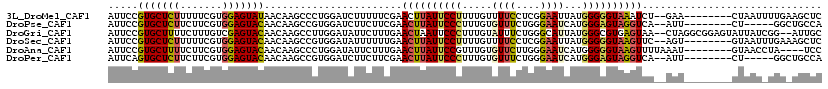
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,058,824 – 16,058,933 |
| Length | 109 |
| Max. P | 0.566665 |

| Location | 16,058,824 – 16,058,933 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.69 |
| Mean single sequence MFE | -27.27 |
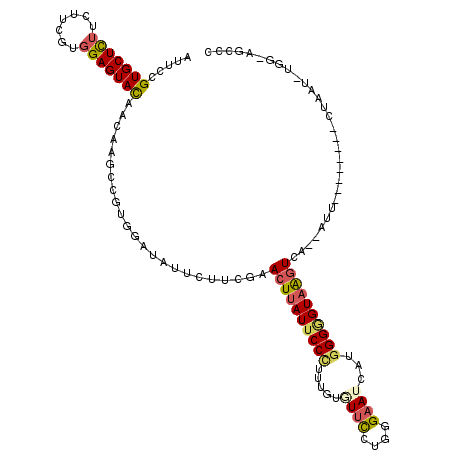
| Consensus MFE | -18.35 |
| Energy contribution | -18.43 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16058824 109 + 23771897 AUUCCGUGCUCUUUUUCGUGGAGUAUAACAAGCCCUGGAUCUUUUUCGAACUUAUUCCUUUUGUUUUCCUCGGAAUUAUGGGGGUAAAUCU--GAA--------CUAAUUUUGAAGCUC .....(((((((.......)))))))....(((..(..(..(..((((.(.((((((((......(((....)))....)))))))).).)--)))--------..)..)..)..))). ( -18.40) >DroPse_CAF1 3607 104 + 1 AUUCCGUGCUCUUCUUCGUGGAGUACAACAAGCCGUGGAUCUUCUUCGAACUUAUUCCCUUUGUGUUUCUGGGAAUCAUGGGAGUAGGUCA--AUU--------CU-----GGCUGCCA .....(((((((.......)))))))....(((((..(((((.((((.(....((((((...........))))))..).)))).))))).--...--------.)-----)))).... ( -30.80) >DroGri_CAF1 8847 115 + 1 AUUCCGUGCUUUUCUUUGUCGAGUACAACAAGCCUUGGAUAUUCUUUGAACUAAUUCCCUUUGUAUUUCUGGGCAUUAUGGGCGUGAGUAA--CUAGGCGGAGUAUUAUCGG--AUUGC (((((((((((.........)))))).....((((.((.(((((..((..(((((.(((...........))).)))).)..)).))))).--)))))))))))........--..... ( -28.50) >DroSec_CAF1 551 109 + 1 AUUCCGUGCUCUUUUUCGUGGAGUACAACAAGCCGUGGAUAUUUUUUGAACUUAUUCCUUUUGUUUUCCUCGGAAUUAUGGGGGUAAGUUC--AGU--------GUAAUUUGAAAGCUC .....(((((((.......)))))))....(((..(.((......((((((((((((((......(((....)))....))))))))))))--)).--------.....)).)..))). ( -31.30) >DroAna_CAF1 813 107 + 1 AUUCCGUGCUUUUCUUCGUGGAGUACAACAAGCCCUGGAUAUUCUUUGAACUUAUUCCGUUUGUGUUCUUGGGAAUCAUGGGGGUAAGUUUUAAAU--------GUAACCUA----UCC .....(((((((.......)))))))..........((.(((.....(((((((((((...((.((((....))))))..))))))))))).....--------))).))..----... ( -23.90) >DroPer_CAF1 3614 104 + 1 AUUCAGUGCUCUUCUUCGUGGAGUACAACAAGCCGUGGAUCUUCUUCGAACUUAUUCCCUUUGUGUUUCUGGGAAUCAUGGGAGUAGGUCA--AUU--------CU-----GGCUGCCA .....(((((((.......)))))))....(((((..(((((.((((.(....((((((...........))))))..).)))).))))).--...--------.)-----)))).... ( -30.70) >consensus AUUCCGUGCUCUUCUUCGUGGAGUACAACAAGCCGUGGAUAUUCUUCGAACUUAUUCCCUUUGUGUUCCUGGGAAUCAUGGGGGUAAGUCA__AUU________CUAAU_UGG_AGCCC .....(((((((.......))))))).......................((((((((((.....((((....))))...)))))))))).............................. (-18.35 = -18.43 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:03 2006