| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,751,065 – 1,751,185 |
| Length | 120 |
| Max. P | 0.535409 |

| Location | 1,751,065 – 1,751,185 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.67 |
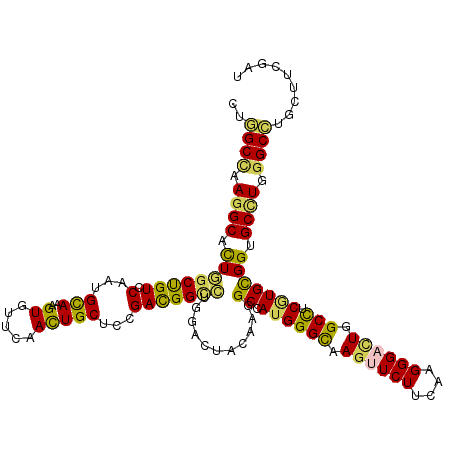
| Mean single sequence MFE | -46.75 |
| Consensus MFE | -29.76 |
| Energy contribution | -28.38 |
| Covariance contribution | -1.38 |
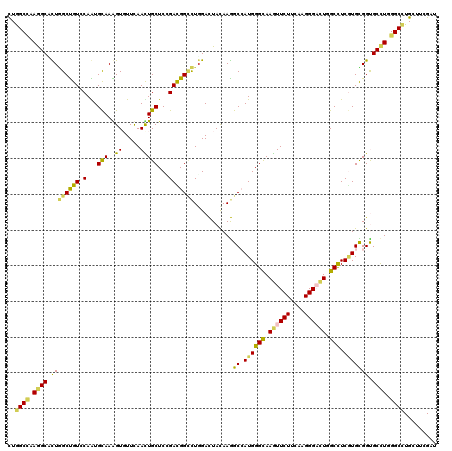
| Combinations/Pair | 1.42 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
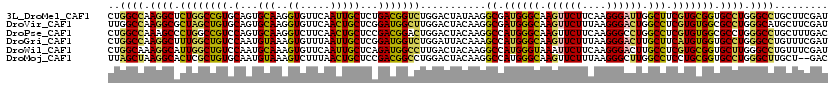
>3L_DroMel_CAF1 1751065 120 - 23771897 CUGGCCAAGGCUCUGGCCGUGCAGUGCAAGGUGUUCAAUUGCUCUGACGGUCUGGACUAUAAGGCGAUGGGCAAGUUCUUCAAGGGAUUGGCUUCGUGCGGUGCCUGGGCCUGCUUCGAU ..((((.((((.((((((((.(((.((((.........)))).)))))))))...........(((..((((.((((((....)))))).))))..))))).)))).))))......... ( -45.60) >DroVir_CAF1 8261 120 - 1 UUGGCCAAGGCGCUAGCUGUGCAGUGCAAGGUGUUCAACUGCUCGGAUGGCUUGGACUACAAGGCGAUGGGCAAGUUCUUUAAGGGACUGGCCUCGUGUGGCGCCUGGGCAUGCUUCGAU ...(((.(((((((((((.((.(((.((((.(((((........))))).)))).))).)).))).((((((.((((((....)))))).))).))).)))))))).))).......... ( -51.50) >DroPse_CAF1 9075 120 - 1 CUGGCCAAAGCCCUGGCCGUCCAGUGCAAGGUCUUCAACUGCUCCGACGGACUGGACUACAAGGCCAUGGGCAAGUUCUUCAAGGGCCUGGCCUCGUGUGGCGCCUGGGCCUGCUUUGAC ..((((...((((((((((((((((.(..((............))...).))))))).....))))).))))...........((((...(((......))))))).))))......... ( -48.20) >DroGri_CAF1 9065 120 - 1 CUGGCCAAGGCUUUGGCUGUCCAAUGUAAAGUGUUUAAUUGCUCGGAUGGUCUGGAUUACAAAGCCAUGGGCAAGUUCUUUAAGGGACUUGCUUCAUGUGGUGCCUGGGCCUGUUUCGAU ..((((.((((...((((((((...((((.........))))..))))))))...........((((((((((((((((....)))))))))))...))))))))).))))......... ( -45.10) >DroWil_CAF1 8224 120 - 1 CUGGCAAAGGCAUUGGCUGUCCAAUGCAAAGUGUUCAAUUGCUCAGAUGGCCUUGACUACAAGGCCAUGGGUAAAUUCUUCAAGGGACUUGCCUCGUGCGGUGCUUGGGCCUGUUUCGAU ..(((((..(((((((....))))))).....((((..(((((((..((((((((....)))))))))))))))...(.....))))))))))(((.((((.((....))))))..))). ( -44.70) >DroMoj_CAF1 8327 118 - 1 UUAGCUAAGGCACUCGCUGUGCAAUGUAAAGUCUUUAACUGCUCCGACGGCCUGGACUACAAGGCCAUGGGCAAGUUCUUUAAGGGCUUGGCCUCCUGCGGUGCCUGGGCUUGCU--GAC (((((....((((.....))))......((((((...(((((......(((((........)))))..((.(((((((.....))))))).))....)))))....)))))))))--)). ( -45.40) >consensus CUGGCCAAGGCACUGGCUGUCCAAUGCAAAGUGUUCAACUGCUCCGACGGCCUGGACUACAAGGCCAUGGGCAAGUUCUUCAAGGGACUGGCCUCGUGCGGUGCCUGGGCCUGCUUCGAU ..((((.((((.((((((((.(...(((..((.....)))))...)))))))...........((.((((((.((((((....)))))).))).))))))).)))).))))......... (-29.76 = -28.38 + -1.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:23 2006