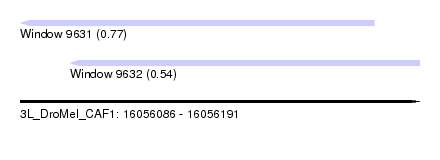
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,056,086 – 16,056,191 |
| Length | 105 |
| Max. P | 0.772119 |

| Location | 16,056,086 – 16,056,179 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 87.67 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -23.55 |
| Energy contribution | -23.13 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16056086 93 - 23771897 ACCUUUUUCAUGACCGUUUGGUAGGCGGUUAUGUAGCUGUUGCCCUCCGCGGCGGCCUCCCUGCUGCCCUUCAGCGGAAACUUCUCCAUCCUG ........((((((((((.....))))))))))..((((..((.....))((((((......))))))...))))(((......)))...... ( -31.00) >DroSec_CAF1 6116 93 - 1 ACCUUUUUCAUGACCGUUUGGUAGGCGGUUAUAUAGCUGUUGCCCUCCGCGGCGGCCUCCCUGCUGCCCUUCAGCGGAAACUUCUCCAUCCUG .............(((((.((..((((((......((((((((.....))))))))......))))))..)))))))................ ( -29.40) >DroEre_CAF1 5195 93 - 1 ACCUUUUUCAUGACCGUUUGGUAGGCGGUUAUGUAGCUGUUGCCCUCCGCGGCGGCCUCCCUGCUGCCCUUCAGCGGAAACUUCUCCAUCCUG ........((((((((((.....))))))))))..((((..((.....))((((((......))))))...))))(((......)))...... ( -31.00) >DroWil_CAF1 2438 93 - 1 ACCUUUUUCAUUACUGUUUGAUAUGCCGUUAUAUAGCUAUUACCCUCUGCAGCAGCCUCUCUGCUUCCUUUUAAUGGGAAUUUUUCCAUUCCU ........(((((((((.((((.....)))).))))..............(((((.....)))))......)))))(((((......))))). ( -12.80) >DroYak_CAF1 5116 93 - 1 ACCUUUUUCAUGACUGUUUGGUAGGCGGUUAUGUAGUUGUUGCCCUCCGCGGCGGCCUCCCUGCUGCCCUUCAGCGGAAACUUCUCCAUCCUG ........((((((((((.....))))))))))............(((((((((((......)))))).....)))))............... ( -26.80) >DroAna_CAF1 4147 89 - 1 ACCUUUUUCAUCACUGUUUGGUAGGCGGUUAUGUAGUUGUUGCCCUCGGCGGCCGCCUCUCGACUGCCCUUCAGCGGAAACUUCUCCAU---- .............(((((.((..(((((((..(..((.((((((...)))))).))..)..)))))))..)))))))............---- ( -26.70) >consensus ACCUUUUUCAUGACCGUUUGGUAGGCGGUUAUGUAGCUGUUGCCCUCCGCGGCGGCCUCCCUGCUGCCCUUCAGCGGAAACUUCUCCAUCCUG .............(((((.((..((((((......((((((((.....))))))))......))))))..)))))))................ (-23.55 = -23.13 + -0.41)



| Location | 16,056,099 – 16,056,191 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 84.91 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -23.55 |
| Energy contribution | -23.59 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16056099 92 - 23771897 -----UUAUGGUGGGCUACCUUUUUCAUGACCGUUUGGUAGGCGGUUAUGUAGCUGUUGCCCUCCGCGGCGGCCUCCCUGCUGCCCUUCAGCGGAAA -----.......((((.((......((((((((((.....)))))))))).....)).))))(((((((((((......)))))).....))))).. ( -38.20) >DroSec_CAF1 6129 92 - 1 -----UUAUGGUGAGCUACCUUUUUCAUGACCGUUUGGUAGGCGGUUAUAUAGCUGUUGCCCUCCGCGGCGGCCUCCCUGCUGCCCUUCAGCGGAAA -----((((((.(((....)))..))))))(((((.((..((((((......((((((((.....))))))))......))))))..)))))))... ( -32.20) >DroSim_CAF1 4643 92 - 1 -----UUAUGGUGAGCUACCUUUUUCAUGACCGUUUGGUAGGCGGUUAUGUAGCUGUUGCCCUCCGCGGCGGCCUCCCUGCUGCCCUUCAGCGGAAA -----....((..((((((.......(((((((((.....))))))))))))))..)..)).(((((((((((......)))))).....))))).. ( -34.81) >DroWil_CAF1 2451 92 - 1 -----AUUUGAUGGCUUACCUUUUUCAUUACUGUUUGAUAUGCCGUUAUAUAGCUAUUACCCUCUGCAGCAGCCUCUCUGCUUCCUUUUAAUGGGAA -----...(((((((.........(((........)))...)))))))....((...........))(((((.....)))))((((......)))). ( -16.00) >DroYak_CAF1 5129 92 - 1 -----UUAUGGUGGGCUACCUUUUUCAUGACUGUUUGGUAGGCGGUUAUGUAGUUGUUGCCCUCCGCGGCGGCCUCCCUGCUGCCCUUCAGCGGAAA -----.......((((.((..((..((((((((((.....)))))))))).))..)).))))(((((((((((......)))))).....))))).. ( -36.00) >DroAna_CAF1 4156 96 - 1 AUCACUUAUGACUC-GUACCUUUUUCAUCACUGUUUGGUAGGCGGUUAUGUAGUUGUUGCCCUCGGCGGCCGCCUCUCGACUGCCCUUCAGCGGAAA ..............-...............(((((.((..(((((((..(..((.((((((...)))))).))..)..)))))))..)))))))... ( -26.70) >consensus _____UUAUGGUGGGCUACCUUUUUCAUGACCGUUUGGUAGGCGGUUAUGUAGCUGUUGCCCUCCGCGGCGGCCUCCCUGCUGCCCUUCAGCGGAAA .....((((((.............))))))(((((.((..((((((......((((((((.....))))))))......))))))..)))))))... (-23.55 = -23.59 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:01 2006