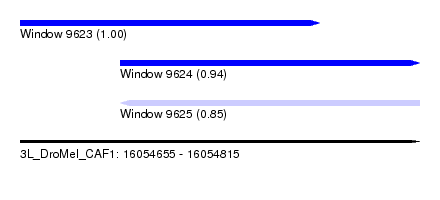
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,054,655 – 16,054,815 |
| Length | 160 |
| Max. P | 0.995432 |

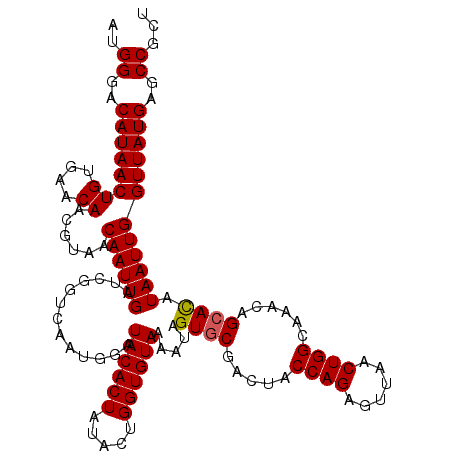
| Location | 16,054,655 – 16,054,775 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.75 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -27.08 |
| Energy contribution | -28.20 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16054655 120 + 23771897 CAUGUGGUGAACAACAUUCCUCUCGUAUUUACCAGACAGCAGCGGCUCAUAACCAAUUAUAUGCUGUUUGCCAGUUAACUCUGGUAGUCGCACUAUUUUACACCAGUAUAGUGUAUCCAU ....(((((((..((.........)).)))))))(((...((((((..((((....))))..))))))((((((......)))))))))(((((((...........)))))))...... ( -28.90) >DroSec_CAF1 4681 120 + 1 CAUGUGGUGAACCACAUUCCUAUCGUAUUUACCAGACAGCAGCGGCUCAUAACCAAUUAUGUGCUGUUUACCAGUUAACUCUGGUAGUCGCACUAUUUUACACCAGUAUAGUGUAUCCAU .((((((....)))))).................(((...((((((.(((((....))))).))))))((((((......)))))))))(((((((...........)))))))...... ( -36.00) >DroSim_CAF1 3195 120 + 1 CAUGUGGUGAACCACAUUCCUUUCGUAUUUACCAGACAGCAGCGGCUCAUAACCAAUUAUGUGCUGUUUACCAGUUAACUCUGGUAGUCGCACUAUUUUACACCAGUAUAGUGUAUCCAU .((((((....)))))).................(((...((((((.(((((....))))).))))))((((((......)))))))))(((((((...........)))))))...... ( -36.00) >DroEre_CAF1 3792 120 + 1 CAUGUGGUGAACAACAUUCGUAUUGUAUUUACCAGACAGCAGAGGCUCAUAACCAAUUAUGUGCUGUUUGCCAGUUAACUCUGGGAGUCGAACUAUUUUACACCAGUAUAGUGUAUCCAU ....(((((((..(((.......))).)))))))(((.((((((((.(((((....))))).))).)))))(((......)))...))).........(((((.......)))))..... ( -27.00) >DroYak_CAF1 3712 120 + 1 CAUGUGGUGAACAACAUUCGUAUUGUAUUUACCAGACAGCAGAGGCUCAUAACCAAUUAUGUGCUGUUUGCCAGUUAACUCUGGGAGUCGCACUAUUUUACACCAGUAUAGUGUAUCCAU ..(.(((((((..(((.......))).))))))).)..((((((((.(((((....))))).))).)))))(((......)))(((...(((((((...........))))))).))).. ( -30.10) >consensus CAUGUGGUGAACAACAUUCCUAUCGUAUUUACCAGACAGCAGCGGCUCAUAACCAAUUAUGUGCUGUUUGCCAGUUAACUCUGGUAGUCGCACUAUUUUACACCAGUAUAGUGUAUCCAU ....(((((((..((.........)).)))))))(((...((((((.(((((....))))).))))))((((((......)))))))))(((((((...........)))))))...... (-27.08 = -28.20 + 1.12)

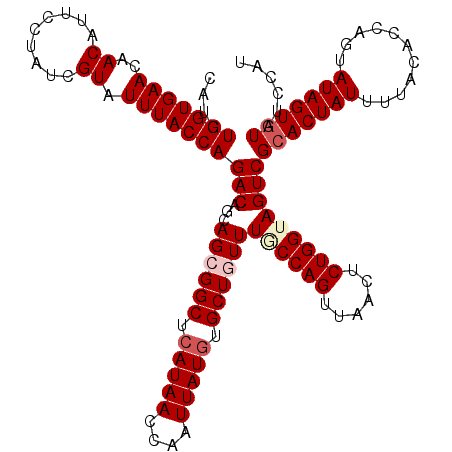
| Location | 16,054,695 – 16,054,815 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -25.50 |
| Energy contribution | -26.06 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16054695 120 + 23771897 AGCGGCUCAUAACCAAUUAUAUGCUGUUUGCCAGUUAACUCUGGUAGUCGCACUAUUUUACACCAGUAUAGUGUAUCCAUUGACCGAUCAAUUGUUACGUUGUUCACAGUUAUGUCCCAU ...((..((((((((((....(((...(((((((......)))))))..)))......(((((.......)))))...))))...((.((((......)))).))...))))))..)).. ( -26.80) >DroSec_CAF1 4721 120 + 1 AGCGGCUCAUAACCAAUUAUGUGCUGUUUACCAGUUAACUCUGGUAGUCGCACUAUUUUACACCAGUAUAGUGUAUCCAUUGAUCGAUCAAUUGUUACGUUGUUCACAGUUAUGUCCCAU ...((..((((((.......((((...(((((((......)))))))..))))............(.((((((((.(.(((((....))))).).)))))))).)...))))))..)).. ( -29.90) >DroSim_CAF1 3235 120 + 1 AGCGGCUCAUAACCAAUUAUGUGCUGUUUACCAGUUAACUCUGGUAGUCGCACUAUUUUACACCAGUAUAGUGUAUCCAUUGACCGAUCAAUUGUUACGUUGUUCACAGUUAUGUCCCAU ...((..((((((((((...((((...(((((((......)))))))..)))).....(((((.......)))))...))))...((.((((......)))).))...))))))..)).. ( -29.90) >DroEre_CAF1 3832 120 + 1 AGAGGCUCAUAACCAAUUAUGUGCUGUUUGCCAGUUAACUCUGGGAGUCGAACUAUUUUACACCAGUAUAGUGUAUCCAUUGACUGCUCAAUUGUUACGUUGUUCACAGUUAUGUCCCAC ...((..((((((......((.((.....))))((.(((..((((((((((.......(((((.......)))))....)))))).))))...)))))..........))))))..)).. ( -25.20) >DroYak_CAF1 3752 120 + 1 AGAGGCUCAUAACCAAUUAUGUGCUGUUUGCCAGUUAACUCUGGGAGUCGCACUAUUUUACACCAGUAUAGUGUAUCCAUUGACUUCUCAAUUGUUACGUUGUUUACAGUUAUGUCCCAU ...((..((((((.......((((...((.((((......)))).))..))))............((((((((((.(.(((((....))))).).))))))))..)).))))))..)).. ( -26.70) >consensus AGCGGCUCAUAACCAAUUAUGUGCUGUUUGCCAGUUAACUCUGGUAGUCGCACUAUUUUACACCAGUAUAGUGUAUCCAUUGACCGAUCAAUUGUUACGUUGUUCACAGUUAUGUCCCAU ...((..((((((.......((((...(((((((......)))))))..))))............(.((((((((.(.(((((....))))).).)))))))).)...))))))..)).. (-25.50 = -26.06 + 0.56)



| Location | 16,054,695 – 16,054,815 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -26.32 |
| Energy contribution | -26.36 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16054695 120 - 23771897 AUGGGACAUAACUGUGAACAACGUAACAAUUGAUCGGUCAAUGGAUACACUAUACUGGUGUAAAAUAGUGCGACUACCAGAGUUAACUGGCAAACAGCAUAUAAUUGGUUAUGAGCCGCU ..((..((((((((....))......((((((.............((((((.....)))))).....((((.....((((......))))......)))).))))))))))))..))... ( -26.20) >DroSec_CAF1 4721 120 - 1 AUGGGACAUAACUGUGAACAACGUAACAAUUGAUCGAUCAAUGGAUACACUAUACUGGUGUAAAAUAGUGCGACUACCAGAGUUAACUGGUAAACAGCACAUAAUUGGUUAUGAGCCGCU ..((..((((((((....))......((((((.............((((((.....)))))).....((((...((((((......))))))....)))).))))))))))))..))... ( -31.60) >DroSim_CAF1 3235 120 - 1 AUGGGACAUAACUGUGAACAACGUAACAAUUGAUCGGUCAAUGGAUACACUAUACUGGUGUAAAAUAGUGCGACUACCAGAGUUAACUGGUAAACAGCACAUAAUUGGUUAUGAGCCGCU ..((..((((((((....))......((((((.............((((((.....)))))).....((((...((((((......))))))....)))).))))))))))))..))... ( -31.60) >DroEre_CAF1 3832 120 - 1 GUGGGACAUAACUGUGAACAACGUAACAAUUGAGCAGUCAAUGGAUACACUAUACUGGUGUAAAAUAGUUCGACUCCCAGAGUUAACUGGCAAACAGCACAUAAUUGGUUAUGAGCCUCU (.((..((((((((....))......((((((.(((((((((...((((((.....)))))).....))).)))).((((......))))......))...))))))))))))..)).). ( -25.60) >DroYak_CAF1 3752 120 - 1 AUGGGACAUAACUGUAAACAACGUAACAAUUGAGAAGUCAAUGGAUACACUAUACUGGUGUAAAAUAGUGCGACUCCCAGAGUUAACUGGCAAACAGCACAUAAUUGGUUAUGAGCCUCU ..(((.((((((((....))......((((((.............((((((.....)))))).....((((.....((((......))))......)))).))))))))))))..))).. ( -26.60) >consensus AUGGGACAUAACUGUGAACAACGUAACAAUUGAUCGGUCAAUGGAUACACUAUACUGGUGUAAAAUAGUGCGACUACCAGAGUUAACUGGCAAACAGCACAUAAUUGGUUAUGAGCCGCU ..((..((((((((....))......((((((.............((((((.....)))))).....((((.....((((......))))......)))).))))))))))))..))... (-26.32 = -26.36 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:54 2006