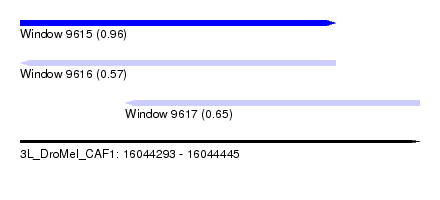
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,044,293 – 16,044,445 |
| Length | 152 |
| Max. P | 0.958047 |

| Location | 16,044,293 – 16,044,413 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -24.36 |
| Consensus MFE | -19.22 |
| Energy contribution | -20.22 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.958047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16044293 120 + 23771897 CGUUAUAUCGUUUCUAUUAGAACUGGAAAAGCAUCGAUGUUUUUGCCAGGCCAACAUCAAUAACCAAUAAUCAUCGAUGUUUUUCUUGCUCCUGGUUCGUUACAAUACAAUACUAAACAC .........((((.((((.((((((((((((((((((((...(((...((.............))..))).))))))))))))))).......)))))((......))))))..)))).. ( -23.43) >DroSec_CAF1 14709 105 + 1 CGUUUUAUCGUUUCUAUUAGAACUAGAAAAGCAUCGAUGUUUUUGCCAGGACUACAUCGAUAACCAAUAAUCAUCGAUAUUUUUCUUGCUCCUGGUUCGUUA---------------CAC ...................((((((((((((((....)))))))((.((((..(.((((((...........)))))).)..)))).))..)))))))....---------------... ( -20.80) >DroSim_CAF1 12215 120 + 1 CGUUUUAUCGUUUCUAUUAGAACUAGAAAAGCAUCGAUGUUUUUGCCAGGACUACAUCGAUAACCAAUAAUCAUCGAUGUUUUUCUUGCUCCUGGUUCGUUACAAUACAAUACUUAACAC .(((.(((.((........((((((((((((((....)))))))((.((((..((((((((...........))))))))..)))).))..)))))))........)).)))...))).. ( -28.19) >DroEre_CAF1 14676 116 + 1 CGUUAUAUCGUUUCUCUUAGAACUGGAAAAUCAUCGAUGUUUUUGC-AGGUAAACAUCGAUAGCCAAUAAUCAUCGAUGUUUUUCUUGCUUCUAUUUCGUUAC--U-CAAUACUUAACAC .((((............((((((.((((((.((((((((...(((.-.(((...........)))..))).)))))))).)))))).).)))))....((...--.-....)).)))).. ( -22.90) >DroYak_CAF1 37150 114 + 1 CGUUAUAUCGUUUCUAUUAGAACUGGAAAAGCAUCGAUGUUUUUGC-AGGUAAACAUCGAUAACCAAUAAUCAUCGAUGUUUUUCUUGCUUCUGUUUCGUUAC--UACAAUACAUAA--- .((..((.((.......((((((.(((((((((((((((...(((.-.(((...........)))..))).))))))))))))))).).)))))...)).)).--.)).........--- ( -26.50) >consensus CGUUAUAUCGUUUCUAUUAGAACUGGAAAAGCAUCGAUGUUUUUGCCAGGACAACAUCGAUAACCAAUAAUCAUCGAUGUUUUUCUUGCUCCUGGUUCGUUAC__UACAAUACUUAACAC ...................((((((((((((((....)))))))((.(((..(((((((((...........)))))))))..))).))..)))))))...................... (-19.22 = -20.22 + 1.00)



| Location | 16,044,293 – 16,044,413 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.66 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16044293 120 - 23771897 GUGUUUAGUAUUGUAUUGUAACGAACCAGGAGCAAGAAAAACAUCGAUGAUUAUUGGUUAUUGAUGUUGGCCUGGCAAAAACAUCGAUGCUUUUCCAGUUCUAAUAGAAACGAUAUAACG ..........(((((((((..(......(((((..(((((.((((((((......(((((.......))))).........)))))))).)))))..)))))....)..))))))))).. ( -28.46) >DroSec_CAF1 14709 105 - 1 GUG---------------UAACGAACCAGGAGCAAGAAAAAUAUCGAUGAUUAUUGGUUAUCGAUGUAGUCCUGGCAAAAACAUCGAUGCUUUUCUAGUUCUAAUAGAAACGAUAAAACG ...---------------..........(((((.((((((.((((((((..(((((.....)))))..((....)).....)))))))).)))))).))))).................. ( -23.20) >DroSim_CAF1 12215 120 - 1 GUGUUAAGUAUUGUAUUGUAACGAACCAGGAGCAAGAAAAACAUCGAUGAUUAUUGGUUAUCGAUGUAGUCCUGGCAAAAACAUCGAUGCUUUUCUAGUUCUAAUAGAAACGAUAAAACG ..(((((((((((...(((......((((((.........(((((((((((.....)))))))))))..)))))).....))).))))))))(((((.......))))).......))). ( -30.30) >DroEre_CAF1 14676 116 - 1 GUGUUAAGUAUUG-A--GUAACGAAAUAGAAGCAAGAAAAACAUCGAUGAUUAUUGGCUAUCGAUGUUUACCU-GCAAAAACAUCGAUGAUUUUCCAGUUCUAAGAGAAACGAUAUAACG ..(((..((((((-.--.(..(....(((((.(..(((((.((((((((.........))))(((((((....-....))))))))))).)))))..)))))).)..)..))))))))). ( -20.60) >DroYak_CAF1 37150 114 - 1 ---UUAUGUAUUGUA--GUAACGAAACAGAAGCAAGAAAAACAUCGAUGAUUAUUGGUUAUCGAUGUUUACCU-GCAAAAACAUCGAUGCUUUUCCAGUUCUAAUAGAAACGAUAUAACG ---...(((((((((--(((.(((.......(((.(..(((((((((((((.....)))))))))))))..))-)).......))).)))).......(((.....)))))))))))... ( -27.84) >consensus GUGUUAAGUAUUGUA__GUAACGAACCAGGAGCAAGAAAAACAUCGAUGAUUAUUGGUUAUCGAUGUUGACCUGGCAAAAACAUCGAUGCUUUUCCAGUUCUAAUAGAAACGAUAUAACG ............................(((((..(((((.((((((((((.....))))))((((((...........)))))))))).)))))..))))).................. (-17.98 = -18.66 + 0.68)


| Location | 16,044,333 – 16,044,445 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 84.89 |
| Mean single sequence MFE | -23.54 |
| Consensus MFE | -16.00 |
| Energy contribution | -16.28 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16044333 112 - 23771897 UAUCCAUAUAAUUUUUUUAACGACAUCGAAGUGUGUUUAGUAUUGUAUUGUAACGAACCAGGAGCAAGAAAAACAUCGAUGAUUAUUGGUUAUUGAUGUUGGCCUGGCAAAA .....(((((((......(((.((((....)))))))....))))))).........(((((.........((((((((((((.....))))))))))))..)))))..... ( -23.70) >DroSec_CAF1 14749 97 - 1 UAUCCAUAUAAUAUGAUUAACGACAUCGAAAUGUG---------------UAACGAACCAGGAGCAAGAAAAAUAUCGAUGAUUAUUGGUUAUCGAUGUAGUCCUGGCAAAA ....(((((....((((.......))))..)))))---------------.......((((((.........(((((((((((.....)))))))))))..))))))..... ( -18.80) >DroSim_CAF1 12255 112 - 1 UAUCCAUAUAAUAUGUUUAACGACAUCGAAGUGUGUUAAGUAUUGUAUUGUAACGAACCAGGAGCAAGAAAAACAUCGAUGAUUAUUGGUUAUCGAUGUAGUCCUGGCAAAA .....(((((((((..(((((.((((....)))))))))))))))))).........((((((.........(((((((((((.....)))))))))))..))))))..... ( -29.00) >DroEre_CAF1 14716 108 - 1 UAUCCAUAUAAUAGGUUUAACGACAUCGAGGUGUGUUAAGUAUUG-A--GUAACGAAAUAGAAGCAAGAAAAACAUCGAUGAUUAUUGGCUAUCGAUGUUUACCU-GCAAAA ........(((((..((((((.((((....)))))))))))))))-.--..............(((.(..(((((((((((.........)))))))))))..))-)).... ( -21.60) >DroYak_CAF1 37190 105 - 1 UAUCCAUAUAAUAUUUUUAACGACAUCGAAG----UUAUGUAUUGUA--GUAACGAAACAGAAGCAAGAAAAACAUCGAUGAUUAUUGGUUAUCGAUGUUUACCU-GCAAAA ......((((((((...((((.........)----))).))))))))--..............(((.(..(((((((((((((.....)))))))))))))..))-)).... ( -24.60) >consensus UAUCCAUAUAAUAUGUUUAACGACAUCGAAGUGUGUUAAGUAUUGUA__GUAACGAACCAGGAGCAAGAAAAACAUCGAUGAUUAUUGGUUAUCGAUGUUGACCUGGCAAAA ....................((....))...................................((.((...((((((((((((.....))))))))))))...)).)).... (-16.00 = -16.28 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:45 2006