
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,044,054 – 16,044,206 |
| Length | 152 |
| Max. P | 0.933344 |

| Location | 16,044,054 – 16,044,172 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.57 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -35.51 |
| Energy contribution | -35.53 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16044054 118 + 23771897 UU--AUUUUUUUUUGUGUGAUUUUAGCUAUUGCAGAAGCUAAAGACUGAGAACGAUCGUCUGCGAGCCGAGAAUCGGGCCCUAACGCGGGUCGUCUCCAAGCUGACCACCUCGGCUCAGA ..--..................((((((........))))))((((.((......))))))..((((((((.....(((((......)))))(((........)))...))))))))... ( -36.40) >DroSec_CAF1 14471 116 + 1 UU--AUUUU--UUUGUGUGAUUUUAGCUAUUGCAGAAGCUAAAGACUGAGAACGAUCGUUUGCGAGCCGAGAAUCGGGCCCUAACGCGGGUCGUCUCCAAGCUGACCACCUCGGCUCAGA ..--.....--.........((((((((........))))))))...(..(((....)))..)((((((((.....(((((......)))))(((........)))...))))))))... ( -35.60) >DroAna_CAF1 10775 118 + 1 UUUUAUUUU--CUUGUGUGAUUUUAGCUAUUGCAGAAGCUAAAGACUGAGAACGACCGUCUGCGAGCCGAGAAUCGGGCCCUCACCCGGGUCGUCUCGAAGCUGACCACCUCGGCUCAGA .......((--((((.((..((((((((........))))))))))))))))...........((((((((...((((......))))(((((.(.....).)))))..))))))))... ( -38.50) >DroSim_CAF1 11978 116 + 1 UU--AUUUU--UUUGUGUGAUUUUAGCUAUUGCAGAAGCUAAAGACUGAGAACGAUCGUCUGCGAGCCGAGAAUCGGGCCCUAACGCGGGUCGUCUCCAAGCUGACCACCUCGGCUCAGA ..--.....--...........((((((........))))))((((.((......))))))..((((((((.....(((((......)))))(((........)))...))))))))... ( -36.40) >DroEre_CAF1 14444 115 + 1 UU--AUUA---UUUGUGUGAUUUUAGCUAUUGCAGAAGCUAAAGACUGAGAACGAUCGUCUGCGAGCCGAGAAUCGGGCCCUUACGCGGGUCGUCUCCAAGCUGACAACCUCGGCUCAGA ..--....---...........((((((........))))))((((.((......))))))..((((((((.....(((((......)))))(((........)))...))))))))... ( -37.30) >DroYak_CAF1 36918 115 + 1 UU--AUUC---UUUGUGUGAUUUUAGCUAUUGCAGAAGCUAAAGACUGAGAACGAUCGUCUGCGAGCCGAGAAUCGGGCCCUCACGCGGGUCGUCUCCAAGCUGACCACCUCGGCUCAGA (.--.(((---((.((....((((((((........)))))))))).)))))..)........((((((((.....(((((......)))))(((........)))...))))))))... ( -37.10) >consensus UU__AUUUU__UUUGUGUGAUUUUAGCUAUUGCAGAAGCUAAAGACUGAGAACGAUCGUCUGCGAGCCGAGAAUCGGGCCCUAACGCGGGUCGUCUCCAAGCUGACCACCUCGGCUCAGA ......................((((((........))))))((((.((......))))))..((((((((.....(((((......)))))(((........)))...))))))))... (-35.51 = -35.53 + 0.03)

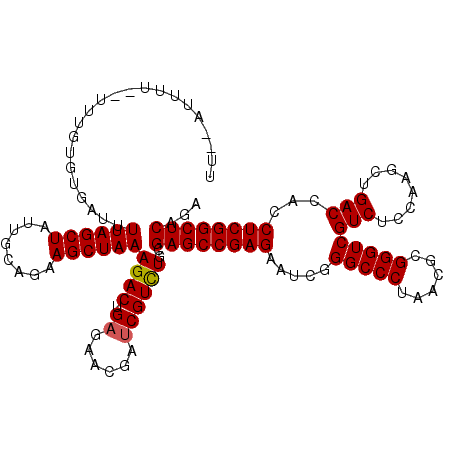

| Location | 16,044,054 – 16,044,172 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.57 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -31.60 |
| Energy contribution | -31.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16044054 118 - 23771897 UCUGAGCCGAGGUGGUCAGCUUGGAGACGACCCGCGUUAGGGCCCGAUUCUCGGCUCGCAGACGAUCGUUCUCAGUCUUUAGCUUCUGCAAUAGCUAAAAUCACACAAAAAAAAAU--AA ...(((((((((.((((.(((((....)))...)).....))))....)))))))))...(((((......)).)))(((((((........))))))).................--.. ( -33.90) >DroSec_CAF1 14471 116 - 1 UCUGAGCCGAGGUGGUCAGCUUGGAGACGACCCGCGUUAGGGCCCGAUUCUCGGCUCGCAAACGAUCGUUCUCAGUCUUUAGCUUCUGCAAUAGCUAAAAUCACACAAA--AAAAU--AA ...(((((((((.((((.(((((....)))...)).....))))....))))))))).................(..(((((((........)))))))..).......--.....--.. ( -32.50) >DroAna_CAF1 10775 118 - 1 UCUGAGCCGAGGUGGUCAGCUUCGAGACGACCCGGGUGAGGGCCCGAUUCUCGGCUCGCAGACGGUCGUUCUCAGUCUUUAGCUUCUGCAAUAGCUAAAAUCACACAAG--AAAAUAAAA .(((((.((((((.....)))))).(((((((.(..((.(((((........))))).))..)))))))))))))..(((((((........)))))))..........--......... ( -38.90) >DroSim_CAF1 11978 116 - 1 UCUGAGCCGAGGUGGUCAGCUUGGAGACGACCCGCGUUAGGGCCCGAUUCUCGGCUCGCAGACGAUCGUUCUCAGUCUUUAGCUUCUGCAAUAGCUAAAAUCACACAAA--AAAAU--AA ...(((((((((.((((.(((((....)))...)).....))))....)))))))))...(((((......)).)))(((((((........)))))))..........--.....--.. ( -33.90) >DroEre_CAF1 14444 115 - 1 UCUGAGCCGAGGUUGUCAGCUUGGAGACGACCCGCGUAAGGGCCCGAUUCUCGGCUCGCAGACGAUCGUUCUCAGUCUUUAGCUUCUGCAAUAGCUAAAAUCACACAAA---UAAU--AA .(((((....(((((((((((.((((.((.(((......)))..)).)))).))))....)))))))...)))))..(((((((........)))))))..........---....--.. ( -34.20) >DroYak_CAF1 36918 115 - 1 UCUGAGCCGAGGUGGUCAGCUUGGAGACGACCCGCGUGAGGGCCCGAUUCUCGGCUCGCAGACGAUCGUUCUCAGUCUUUAGCUUCUGCAAUAGCUAAAAUCACACAAA---GAAU--AA ((((((((((((.((((...(((....)))....(....)))))....)))))))))...(((((......)).)))(((((((........))))))).........)---))..--.. ( -34.60) >consensus UCUGAGCCGAGGUGGUCAGCUUGGAGACGACCCGCGUUAGGGCCCGAUUCUCGGCUCGCAGACGAUCGUUCUCAGUCUUUAGCUUCUGCAAUAGCUAAAAUCACACAAA__AAAAU__AA ...(((((((((..(((...(((....)))(((......)))...))))))))))))...(((((......)).)))(((((((........)))))))..................... (-31.60 = -31.80 + 0.20)

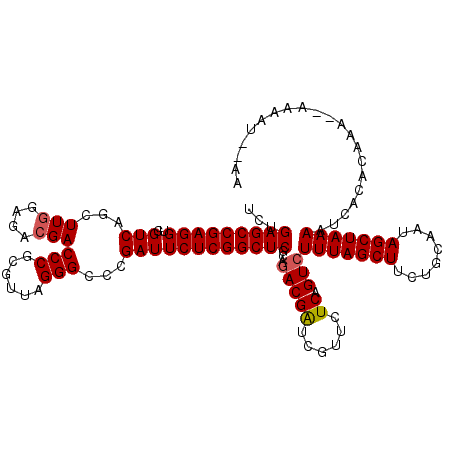
| Location | 16,044,092 – 16,044,206 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.13 |
| Mean single sequence MFE | -40.21 |
| Consensus MFE | -36.26 |
| Energy contribution | -36.32 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16044092 114 + 23771897 AAAGACUGAGAACGAUCGUCUGCGAGCCGAGAAUCGGGCCCUAACGCGGGUCGUCUCCAAGCUGACCACCUCGGCUCAGAGUCAGCUGGCCAAGAGCAAAUAG------GCGCAGAUCAG .....((((((....)).(((((((((((((.....(((((......)))))(((........)))...)))))))).......(((.((.....)).....)------))))))))))) ( -40.80) >DroSec_CAF1 14507 114 + 1 AAAGACUGAGAACGAUCGUUUGCGAGCCGAGAAUCGGGCCCUAACGCGGGUCGUCUCCAAGCUGACCACCUCGGCUCAGAGUCAGCUGGCCAAGACCAAAUAG------GCGCAGAUCAG ...(((((..(((....)))..)((((((((.....(((((......)))))(((........)))...))))))))..))))..((((((...........)------)).)))..... ( -40.00) >DroAna_CAF1 10813 120 + 1 AAAGACUGAGAACGACCGUCUGCGAGCCGAGAAUCGGGCCCUCACCCGGGUCGUCUCGAAGCUGACCACCUCGGCUCAGAGUCAGCUGGCCAAGACCAAAUAGAUCAGAGCUGAGACCCC .....((((........((((..((((((((...((((......))))(((((.(.....).)))))..)))))))).(.(((....)))).))))........))))............ ( -38.79) >DroSim_CAF1 12014 114 + 1 AAAGACUGAGAACGAUCGUCUGCGAGCCGAGAAUCGGGCCCUAACGCGGGUCGUCUCCAAGCUGACCACCUCGGCUCAGAGUCAGCUGGCCAAGACCAAAUAG------GCGCAGAUCAG .....((((((....)).(((((((((((((.....(((((......)))))(((........)))...))))))))...((((..(((......)))..).)------))))))))))) ( -41.00) >DroEre_CAF1 14479 113 + 1 AAAGACUGAGAACGAUCGUCUGCGAGCCGAGAAUCGGGCCCUUACGCGGGUCGUCUCCAAGCUGACAACCUCGGCUCAGAGUCAGCUGGCCAAGACCAAAUAG------GCGCAGAUCA- .................((((((((((((((.....(((((......)))))(((........)))...))))))))...((((..(((......)))..).)------))))))))..- ( -40.80) >DroYak_CAF1 36953 113 + 1 AAAGACUGAGAACGAUCGUCUGCGAGCCGAGAAUCGGGCCCUCACGCGGGUCGUCUCCAAGCUGACCACCUCGGCUCAGAGUCAGCUGGCCAAGACCAAAUAG------GCGCAGAUCA- .................((((((((((((((.....(((((......)))))(((........)))...))))))))...((((..(((......)))..).)------))))))))..- ( -39.90) >consensus AAAGACUGAGAACGAUCGUCUGCGAGCCGAGAAUCGGGCCCUAACGCGGGUCGUCUCCAAGCUGACCACCUCGGCUCAGAGUCAGCUGGCCAAGACCAAAUAG______GCGCAGAUCAG .................((((((((((((((.....(((((......)))))(((........)))...)))))))).(.(((....))))....................))))))... (-36.26 = -36.32 + 0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:42 2006