
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,024,988 – 16,025,128 |
| Length | 140 |
| Max. P | 0.825499 |

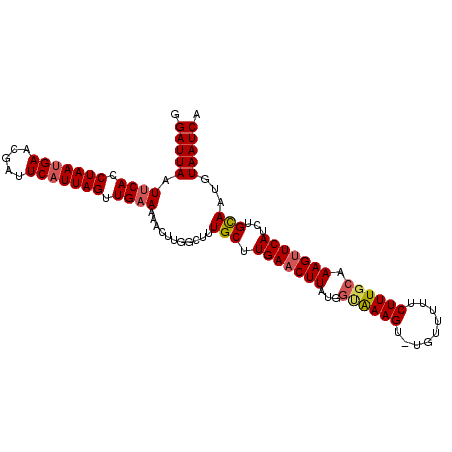
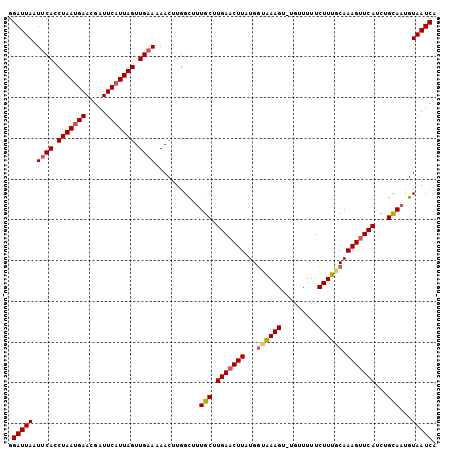
| Location | 16,024,988 – 16,025,088 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -19.75 |
| Energy contribution | -20.87 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16024988 100 + 23771897 GGAUUA-UCCACCUAAGGAACGAUUCAUUAGUUGAACAACUUGGCUUUGCUUGAACUUAUGGUAAAGG----UUUUCUUUCCAAAGCUCAUCUGUAUUGUAAUCA .(((((-(.((.((((.((.....)).)))).)).(((...((((((((...((((((.......)))----)))......)))))).))..)))...)))))). ( -17.30) >DroSec_CAF1 24714 105 + 1 GGAUUAAUUCACCUAAUGAACGAUUCAUUAGUUGAAAAACUUGGCUUUGCUUGAACUUAUGACAAAGUUUGUUUUUCUUUGCAAAGUUCAUCUGCAAUGUAAUCA .((((..((((.(((((((.....))))))).)))).......((.((((.(((((((.((.(((((.........))))))))))))))...)))).)))))). ( -28.30) >DroSim_CAF1 33375 104 + 1 GGAUUAAUUCACCUAAUGAACGAUUCAUUAGUUGAAAAACUUGGUUUUGCUUGAACUUGUGGUGAAGU-UGUUUUUCUUUGCAAAGUUCAUCUGCAAUGUAAUCG .(((((.((((.(((((((.....))))))).))))..........((((.(((((((...(..(((.-.......)))..).)))))))...))))..))))). ( -29.70) >consensus GGAUUAAUUCACCUAAUGAACGAUUCAUUAGUUGAAAAACUUGGCUUUGCUUGAACUUAUGGUAAAGU_UGUUUUUCUUUGCAAAGUUCAUCUGCAAUGUAAUCA .(((((.((((.(((((((.....))))))).))))...........(((.(((((((...((((((.........)))))).)))))))...)))...))))). (-19.75 = -20.87 + 1.12)

| Location | 16,024,988 – 16,025,088 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -15.46 |
| Energy contribution | -15.13 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16024988 100 - 23771897 UGAUUACAAUACAGAUGAGCUUUGGAAAGAAAA----CCUUUACCAUAAGUUCAAGCAAAGCCAAGUUGUUCAACUAAUGAAUCGUUCCUUAGGUGGA-UAAUCC ...............((((((((((((((....----.)))).))).)))))))...........((((((((.((((.((.....)).)))).))))-)))).. ( -20.80) >DroSec_CAF1 24714 105 - 1 UGAUUACAUUGCAGAUGAACUUUGCAAAGAAAAACAAACUUUGUCAUAAGUUCAAGCAAAGCCAAGUUUUUCAACUAAUGAAUCGUUCAUUAGGUGAAUUAAUCC .(((((..((((...(((((((.((((((.........))))))...))))))).))))..........((((.(((((((.....))))))).)))).))))). ( -25.30) >DroSim_CAF1 33375 104 - 1 CGAUUACAUUGCAGAUGAACUUUGCAAAGAAAAACA-ACUUCACCACAAGUUCAAGCAAAACCAAGUUUUUCAACUAAUGAAUCGUUCAUUAGGUGAAUUAAUCC .(((((..((((...(((((((((..(((.......-.)))...)).))))))).))))..........((((.(((((((.....))))))).)))).))))). ( -20.50) >consensus UGAUUACAUUGCAGAUGAACUUUGCAAAGAAAAACA_ACUUUACCAUAAGUUCAAGCAAAGCCAAGUUUUUCAACUAAUGAAUCGUUCAUUAGGUGAAUUAAUCC .(((((.........(((((((......(((........))).....)))))))...............((((.(((((((.....))))))).)))).))))). (-15.46 = -15.13 + -0.33)

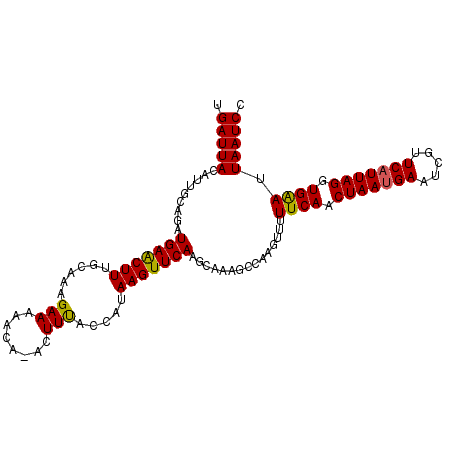
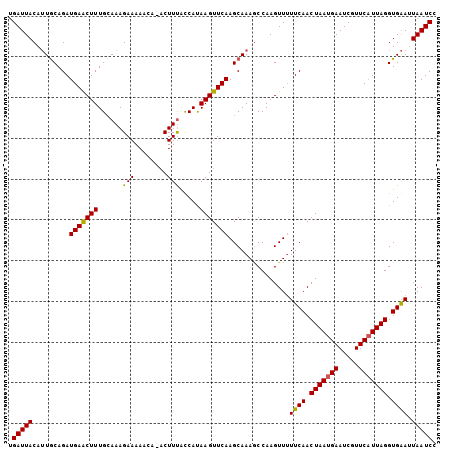
| Location | 16,025,027 – 16,025,128 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.63 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -10.44 |
| Energy contribution | -11.06 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16025027 101 - 23771897 AGCUUUACUAGCGCAGGCUAUUAAAUAUUCAAGUCAUAACUGAUUACAAUACAGAUGAGCUUUGGAAAGAAAA-------------------CCUUUACCAUAAGUUCAAGCAAAGCCAA .(((.....)))...((((............(((((....)))))..........((((((((((((((....-------------------.)))).))).))))))).....)))).. ( -23.00) >DroSec_CAF1 24754 103 - 1 GGCUUAACUAGCGCAGGCUAUUAAAUAUUCAAGUC--AACUGAUUACAUUGCAGAUGAACUUUGCAAAGAAAAACA---------------AACUUUGUCAUAAGUUCAAGCAAAGCCAA .(((.....)))...((((............((((--....))))...((((...(((((((.((((((.......---------------..))))))...))))))).)))))))).. ( -24.10) >DroSim_CAF1 33415 104 - 1 GGCUUAACUAGCACAGGCUAUUAAAUAUUCAAGUCAUAACCGAUUACAUUGCAGAUGAACUUUGCAAAGAAAAACA----------------ACUUCACCACAAGUUCAAGCAAAACCAA .(((((((((((....)))........(((.((((......))))...(((((((.....))))))).))).....----------------...........)))).))))........ ( -16.30) >DroEre_CAF1 27799 115 - 1 AGCUGAACUAGUAC--GCUAUUUCAUAUUCAGGUCAUAACUGAUUAAGUUAGAGAUGAACUUUGGAAUUACAAACAACGCUUACACUAACAAACUUUACUACAAGUUUAAACUAAGC--- .(((......(((.--((.((((((..((((..((.(((((.....)))))))..))))...))))))..........)).)))......((((((......))))))......)))--- ( -17.40) >consensus AGCUUAACUAGCACAGGCUAUUAAAUAUUCAAGUCAUAACUGAUUACAUUACAGAUGAACUUUGCAAAGAAAAACA________________ACUUUACCACAAGUUCAAGCAAAGCCAA .(((.....)))...((((............((((......))))..........((((((((((((((........................)))).))).))))))).....)))).. (-10.44 = -11.06 + 0.62)

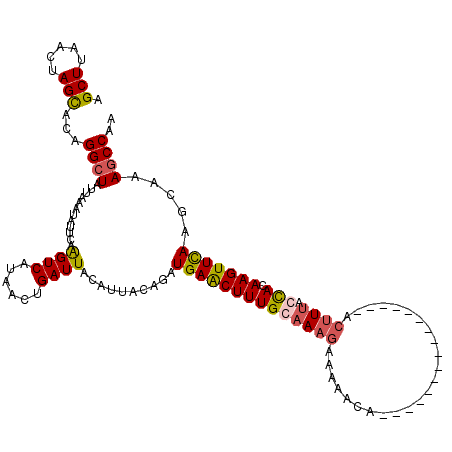
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:32 2006