| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
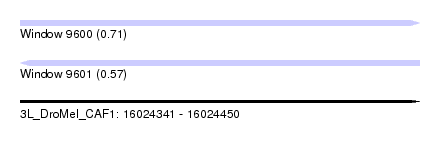
| Location | 16,024,341 – 16,024,450 |
| Length | 109 |
| Max. P | 0.706963 |

| Location | 16,024,341 – 16,024,450 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 90.98 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -16.92 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
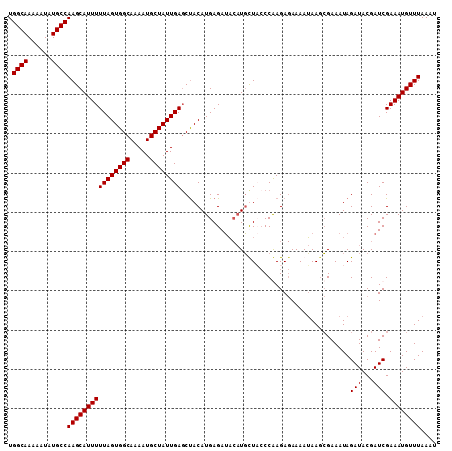
>3L_DroMel_CAF1 16024341 109 + 23771897 UGGCAAAAAUAUGCCAAGCAUUUUUAGUGGCAAAAUGCUAUUGACCUACAUGAGAUACAUGUUCCCCAAGAGAAAAUAAGCGAAAUAGAUACGAUCGAAAUGUUUAAAA .((((......))))((((((((((((((((.....)))))))....(((((.....)))))..................................))))))))).... ( -20.50) >DroSec_CAF1 24127 109 + 1 UGGCAAAAAUAUGCCAAGCAUUUUUAGUGGCAAAAUGCUAUUGAGCUACAUGAGAUACAUGCUACCCGAGAGAAAAUAAUCGAAAUAGAUACGAUCGAAAUGUUUAAAU .((((......))))((((((((((((((((.....)))))))).........(((.....(((..(((..........)))...))).....))).)))))))).... ( -22.00) >DroSim_CAF1 32761 109 + 1 UGGCAAAAAUAUGCCAAGCAUUUUUAGUGGCAAAAUGCUAUUGAGCUACAUGAGAUACAUGCUACCCGAGAGAAAAUAAGCGAAAUAGAUACGAUCGAAAUGUUUAAAU .((((......))))((((((((((((((((.....))))))))(((.((((.....)))).....(....)......)))................)))))))).... ( -22.80) >DroEre_CAF1 27131 104 + 1 UGGCAAAAAUAUGCCAAGCAUUUUUAGUGGCACAAUGCUAUUGAGCCACAUGAGAAC--UUUUCGCCAAGA-AAAAGAAGAGAAAUAGAUACGAUCGAAAUGUUUAA-- .((((......))))(((((((((..(((((.((((...)))).))))).......(--(((((.......-....))))))..............)))))))))..-- ( -26.40) >consensus UGGCAAAAAUAUGCCAAGCAUUUUUAGUGGCAAAAUGCUAUUGAGCUACAUGAGAUACAUGCUACCCAAGAGAAAAUAAGCGAAAUAGAUACGAUCGAAAUGUUUAAAU .((((......))))((((((((((((((((.....))))))))...........................................(((...))).)))))))).... (-16.92 = -16.93 + 0.00)

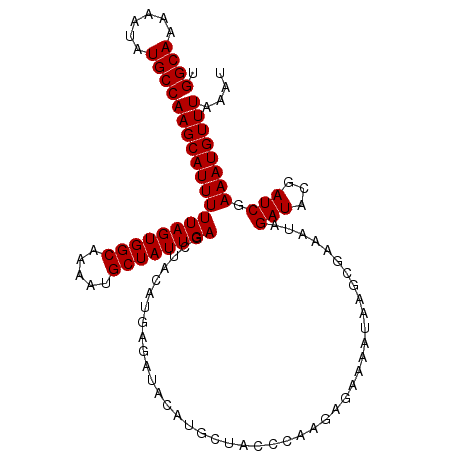
| Location | 16,024,341 – 16,024,450 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 90.98 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -16.05 |
| Energy contribution | -17.80 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16024341 109 - 23771897 UUUUAAACAUUUCGAUCGUAUCUAUUUCGCUUAUUUUCUCUUGGGGAACAUGUAUCUCAUGUAGGUCAAUAGCAUUUUGCCACUAAAAAUGCUUGGCAUAUUUUUGCCA .............((((................((..(....)..))(((((.....))))).))))...((((((((.......))))))))(((((......))))) ( -23.30) >DroSec_CAF1 24127 109 - 1 AUUUAAACAUUUCGAUCGUAUCUAUUUCGAUUAUUUUCUCUCGGGUAGCAUGUAUCUCAUGUAGCUCAAUAGCAUUUUGCCACUAAAAAUGCUUGGCAUAUUUUUGCCA .............(((((.........)))))..........((((.(((((.....))))).))))...((((((((.......))))))))(((((......))))) ( -25.90) >DroSim_CAF1 32761 109 - 1 AUUUAAACAUUUCGAUCGUAUCUAUUUCGCUUAUUUUCUCUCGGGUAGCAUGUAUCUCAUGUAGCUCAAUAGCAUUUUGCCACUAAAAAUGCUUGGCAUAUUUUUGCCA ..........................................((((.(((((.....))))).))))...((((((((.......))))))))(((((......))))) ( -23.10) >DroEre_CAF1 27131 104 - 1 --UUAAACAUUUCGAUCGUAUCUAUUUCUCUUCUUUU-UCUUGGCGAAAA--GUUCUCAUGUGGCUCAAUAGCAUUGUGCCACUAAAAAUGCUUGGCAUAUUUUUGCCA --...................................-...(((((((((--........(((((.((((...)))).))))).....((((...)))).))))))))) ( -21.70) >consensus AUUUAAACAUUUCGAUCGUAUCUAUUUCGCUUAUUUUCUCUCGGGGAACAUGUAUCUCAUGUAGCUCAAUAGCAUUUUGCCACUAAAAAUGCUUGGCAUAUUUUUGCCA ..........................................((((.(((((.....))))).))))...((((((((.......))))))))(((((......))))) (-16.05 = -17.80 + 1.75)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:29 2006