
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,024,133 – 16,024,266 |
| Length | 133 |
| Max. P | 0.906406 |

| Location | 16,024,133 – 16,024,233 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -20.87 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16024133 100 - 23771897 UCCACCAGGUGCGGCCAGACAUCUGGCUGGACUAAAGAGAGAAGUGUGAAAAAUCAUGAACACAAGCUAUUCCACAGUGAAA-------AAUAAACAAGCCGAAAGA ..(((..(((.(((((((....))))))).)))...(((((..((((............))))...)).)))....)))...-------...........(....). ( -23.40) >DroSec_CAF1 23909 100 - 1 UCCACCAGGUGCGGCCAGACAUCUGGCUGGACUAAAGAGAGAAGUGUGAAAAAUCAUGAACACCAGCUAUUCCACAAUGAAA-------AAUAAACAAACCGAAAGA .......(((.(((((((....))))))).)))...(((((..((((............))))...)).)))..........-------...........(....). ( -21.30) >DroSim_CAF1 32536 107 - 1 UCCACCAGGUGCGGCCAGACAUCUGGCUGGACUAAAGCUAGAAGUGUGAAAAAUCAUGAACACCAGCUAUUCCACAGUGAAAAAAAAAAAAUAAACAAACCGAAAGA ..(((..(((.(((((((....))))))).)))..((((....((((............)))).))))........))).....................(....). ( -25.10) >consensus UCCACCAGGUGCGGCCAGACAUCUGGCUGGACUAAAGAGAGAAGUGUGAAAAAUCAUGAACACCAGCUAUUCCACAGUGAAA_______AAUAAACAAACCGAAAGA ..(((..(((.(((((((....))))))).)))......((..((((............))))...))........))).....................(....). (-20.87 = -21.20 + 0.33)

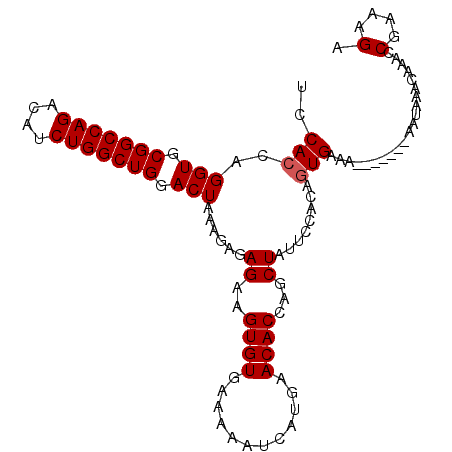
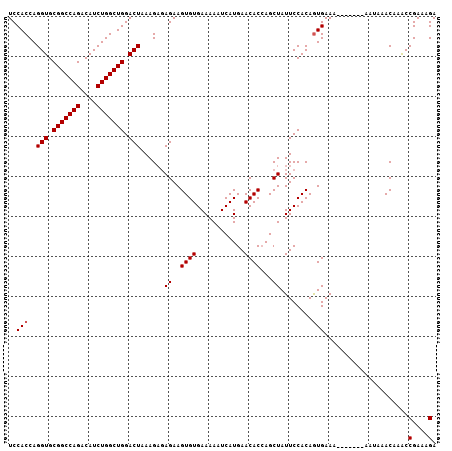
| Location | 16,024,161 – 16,024,266 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 92.86 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -25.80 |
| Energy contribution | -27.13 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16024161 105 - 23771897 AAUAUAUUUUUAUUUCUAAUUUCUUUCGACUAG-------UCCACCAGGUGCGGCCAGACAUCUGGCUGGACUAAAGAGAGAAGUGUGAAAAAUCAUGAACACAAGCUAUUC ............((((..(((((((((...(((-------((((((((((((.....).))))))).)))))))..)))))))))..))))..................... ( -33.20) >DroSec_CAF1 23937 112 - 1 AAUGUAUUUUUAUUUCUAAUUUCUUUCGAGUAGUCAGUCGUCCACCAGGUGCGGCCAGACAUCUGGCUGGACUAAAGAGAGAAGUGUGAAAAAUCAUGAACACCAGCUAUUC .(((.((((((.......(((((((((...(((((......((....))..(((((((....))))))))))))..)))))))))...)))))))))............... ( -31.60) >DroSim_CAF1 32571 112 - 1 AAUAUAUUUUUAUUUCUAAUUUCUUUCGAGUAGUCAGUCGUCCACCAGGUGCGGCCAGACAUCUGGCUGGACUAAAGCUAGAAGUGUGAAAAAUCAUGAACACCAGCUAUUC ...........................(((((((.....(((((((((((((.....).))))))).)))))...........((((............))))..))))))) ( -26.70) >consensus AAUAUAUUUUUAUUUCUAAUUUCUUUCGAGUAGUCAGUCGUCCACCAGGUGCGGCCAGACAUCUGGCUGGACUAAAGAGAGAAGUGUGAAAAAUCAUGAACACCAGCUAUUC ............((((..(((((((((...(((((......((....))..(((((((....))))))))))))..)))))))))..))))..................... (-25.80 = -27.13 + 1.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:27 2006