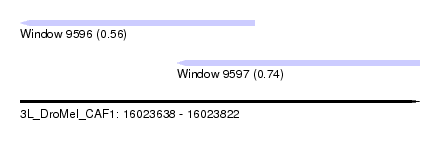
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,023,638 – 16,023,822 |
| Length | 184 |
| Max. P | 0.743840 |

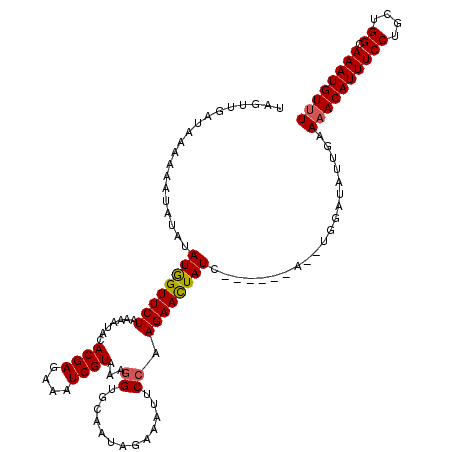
| Location | 16,023,638 – 16,023,746 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 88.03 |
| Mean single sequence MFE | -15.48 |
| Consensus MFE | -11.00 |
| Energy contribution | -11.38 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16023638 108 - 23771897 --A--UGGAUAUUGAAAACAUUUCCUGCUGGCAAAUGUUUAAAAGUUACUACAAUUUCAACAAUAUUUUCCAAGAACAAAAAAUACAACAAUAAUUUCAUUAAACACGUACA --.--(((((((((.((((((((((....)).))))))))....((....))........)))))...))))........................................ ( -13.70) >DroSec_CAF1 23437 108 - 1 --A--UGGAUAUUGAAAACAUUUCCUGCUGGCAAAUGUUUAAAAGUUACUACAAUUUCAACGAUAUGUUCCAAAAACAAAAAAUACAACAAUAAUUUCAUUAAACACAUAUA --.--(((((((((.((((((((((....)).))))))))....((....))........)))))...))))........................................ ( -13.40) >DroSim_CAF1 32052 108 - 1 --A--UGGAUAUUGAAAACAUUUCCUGCUGGCAAAUGUUUAAAAGUUACUACAAUUUCAACAAUAUUUGCCAAGAACAAAAAAUACAACAAUAAUUUCAUUAAACACGUAUA --.--((..((.(((((.((.....)).(((((((((((...(((((.....)))))....)))))))))))......................))))).))..))...... ( -14.30) >DroEre_CAF1 26456 106 - 1 UGAUUUGGAUAUGGAAAACAUUUCCUGUUGGGAAAUGU-UAGAAGUUAGUACGAUUUGAACGAUAUUUUCCAAGAACAAA-----UAUCAAUAAUUCCAUUAAACACGUAUA ..((.((...(((((((((((((((.....))))))))-).....................(((((((.........)))-----)))).....))))))....)).))... ( -20.50) >consensus __A__UGGAUAUUGAAAACAUUUCCUGCUGGCAAAUGUUUAAAAGUUACUACAAUUUCAACAAUAUUUUCCAAGAACAAAAAAUACAACAAUAAUUUCAUUAAACACGUAUA .....(((((((((.((((((((((....)).))))))))....((....))........))))))...)))........................................ (-11.00 = -11.38 + 0.38)

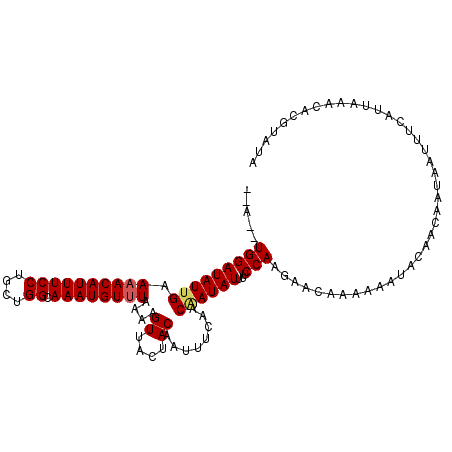
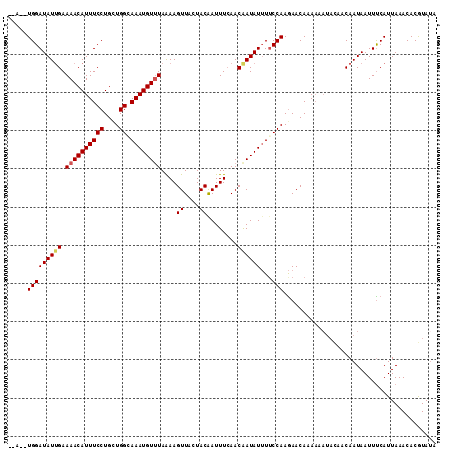
| Location | 16,023,710 – 16,023,822 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -16.59 |
| Energy contribution | -16.72 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16023710 112 - 23771897 UAGUUGAUAAAAAAAAUAUAUGGUUCUAAAAUACACGAGAAAUCGUAAAGGUGCAAUAGAAAUUCCAAGAACUAUC------A--UGGAUAUUGAAAACAUUUCCUGCUGGCAAAUGUUU ...(..(((..............(((((...(((((((....))))....)))...)))))..((((.((....))------.--)))))))..)((((((((((....)).)))))))) ( -20.40) >DroSec_CAF1 23509 112 - 1 UAGUUGAUAAAAAAUAUAUAUGGUUCUAAAAUACACGAGAAAUCGUAAAGGUGCCAUAGAAAUUCCAAGAACUAUG------A--UGGAUAUUGAAAACAUUUCCUGCUGGCAAAUGUUU ...(..(((.....(((.(((((((((.......((((....))))...((.............)).)))))))))------)--))..)))..)((((((((((....)).)))))))) ( -22.12) >DroSim_CAF1 32124 112 - 1 UAGUUGAUAAAAAAUAUAUAUGGUUCUAAAAUACACGAAAAAUCGUAAAGGUGCAAUAGAAAUUCCAAGAACUAUG------A--UGGAUAUUGAAAACAUUUCCUGCUGGCAAAUGUUU ...(..(((.....(((.(((((((((.......((((....))))...((.............)).)))))))))------)--))..)))..)((((((((((....)).)))))))) ( -19.32) >DroEre_CAF1 26523 117 - 1 UCGUUGAUAAAAAAUG--UAUAGUUCUAAAACACACGAGAAAUCGUAAAUGUGUAAUAGAAGUUCCUAGAAUUAUCUAAAUGAUUUGGAUAUGGAAAACAUUUCCUGUUGGGAAAUGU-U ................--.....(((((..((((((((....))))....))))..))))).((((......(((((((.....))))))).))))(((((((((.....))))))))-) ( -27.70) >consensus UAGUUGAUAAAAAAUAUAUAUGGUUCUAAAAUACACGAGAAAUCGUAAAGGUGCAAUAGAAAUUCCAAGAACUAUC______A__UGGAUAUUGAAAACAUUUCCUGCUGGCAAAUGUUU ...................((((((((.......((((....))))...((.............)).))))))))....................((((((((((....)).)))))))) (-16.59 = -16.72 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:26 2006