| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,014,009 – 16,014,103 |
| Length | 94 |
| Max. P | 0.892363 |

| Location | 16,014,009 – 16,014,103 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 60.95 |
| Mean single sequence MFE | -19.38 |
| Consensus MFE | -9.16 |
| Energy contribution | -9.04 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16014009 94 + 23771897 ACUUGAGUCACUUUGUUGCUUUAAUAAUGAAACCAUGCCAGCAUUAACAAGUAUGACUCAAUAGGGAUAUACCUUAAUU-UUAUCUAUAUAUGCA ..((((((((.(((((((((......(((....)))...))))...)))))..))))))))(((((.....)))))...-............... ( -17.90) >DroPse_CAF1 32049 92 + 1 UCUACGGCCUGUGGAUCUAUUGCACAGGCAGUACAGG---UACAAGAGUGUACAAACUUCGACAUUAAAGAUUGCUCACCAGGGCAGUGUUUGUA ..(((.(((((((.(.....).))))))).)))...(---((((....)))))............((((.(((((((....))))))).)))).. ( -29.60) >DroSec_CAF1 13886 92 + 1 ACUUGAGUCACUUUGUUGCUUUAAUAAUGAAACCAUU---GCACUAACAAGUAUGACUCAAUAGGAAUAUGCCUUAAUUUUUAUCUAUAUAUGCA ..((((((((.(((((((......(((((....))))---)...)))))))..)))))))).(((......)))..................... ( -14.90) >DroSim_CAF1 22086 94 + 1 ACUUGAGUCACUUUGUUGCUUUAAUAAUGAAACCAUGCCAGCACUAACAACUAUGACUCAAUAGGAAUAUACCUUAAUU-UUAUCUAUAUAUGCA ..((((((((..((((((((......(((....)))...))))...))))...)))))))).(((......))).....-............... ( -15.10) >consensus ACUUGAGUCACUUUGUUGCUUUAAUAAUGAAACCAUG___GCACUAACAAGUAUGACUCAAUAGGAAUAUACCUUAAUU_UUAUCUAUAUAUGCA ..((((((((..((((((........(((....)))........))))))...)))))))).(((......)))..................... ( -9.16 = -9.04 + -0.12)

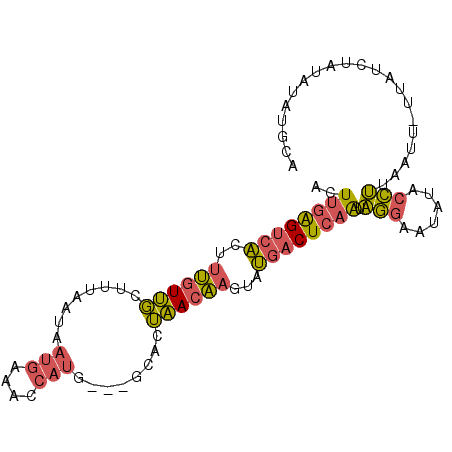
| Location | 16,014,009 – 16,014,103 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 60.95 |
| Mean single sequence MFE | -19.20 |
| Consensus MFE | -9.98 |
| Energy contribution | -8.86 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16014009 94 - 23771897 UGCAUAUAUAGAUAA-AAUUAAGGUAUAUCCCUAUUGAGUCAUACUUGUUAAUGCUGGCAUGGUUUCAUUAUUAAAGCAACAAAGUGACUCAAGU ...............-.....(((......))).(((((((((..((((...((((...(((((...)))))...)))))))).))))))))).. ( -18.10) >DroPse_CAF1 32049 92 - 1 UACAAACACUGCCCUGGUGAGCAAUCUUUAAUGUCGAAGUUUGUACACUCUUGUA---CCUGUACUGCCUGUGCAAUAGAUCCACAGGCCGUAGA .........(((...((((.((((.((((......)))).)))).))))...)))---....(((.(((((((.........))))))).))).. ( -23.00) >DroSec_CAF1 13886 92 - 1 UGCAUAUAUAGAUAAAAAUUAAGGCAUAUUCCUAUUGAGUCAUACUUGUUAGUGC---AAUGGUUUCAUUAUUAAAGCAACAAAGUGACUCAAGU .....................(((......))).(((((((((..(((((.((..---((((((...))))))...))))))).))))))))).. ( -16.50) >DroSim_CAF1 22086 94 - 1 UGCAUAUAUAGAUAA-AAUUAAGGUAUAUUCCUAUUGAGUCAUAGUUGUUAGUGCUGGCAUGGUUUCAUUAUUAAAGCAACAAAGUGACUCAAGU ...............-.....(((......))).(((((((((.((((((((((.(((.......))).)))))..)))))...))))))))).. ( -19.20) >consensus UGCAUAUAUAGAUAA_AAUUAAGGUAUAUUCCUAUUGAGUCAUACUUGUUAGUGC___CAUGGUUUCAUUAUUAAAGCAACAAAGUGACUCAAGU .....................(((......))).(((((((((..(((((...(((...................)))))))).))))))))).. ( -9.98 = -8.86 + -1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:17 2006