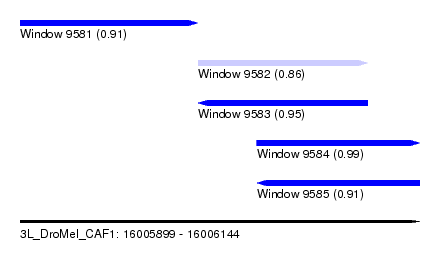
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 16,005,899 – 16,006,144 |
| Length | 245 |
| Max. P | 0.987079 |

| Location | 16,005,899 – 16,006,008 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -18.09 |
| Energy contribution | -18.29 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
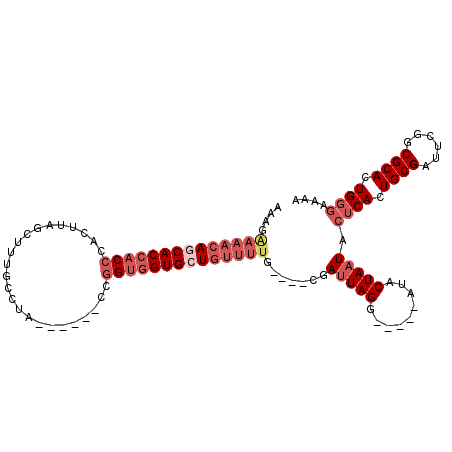
>3L_DroMel_CAF1 16005899 109 + 23771897 GCUAAGGGUGGUUUUUCCCUUC----GGGAAAAACUCCCGAG--UUGCGACUACAAAAAAAAAAAAAAAACAAAUAUUCGUAGUGCCAUCGAAUACGCAAUGUUACAACCGCUAC ((...(((.((((((((((...----)))))))))))))..(--(((((.........................((((((.........)))))))))))).........))... ( -26.41) >DroSec_CAF1 6215 98 + 1 GCCAAGGGUGGUUUUUCCUUUC----GGGAAAAACUCCCGAG--UUGCGACUACAAAACAA-----------AAUAUUCGUAGUGCCAUCGAGUACGCAAUGUUACAACCGCUCG .....((((((((.......((----((((.....))))))(--((((((((((.......-----------.......)))))((......)).)))))).....)))))))). ( -29.54) >DroSim_CAF1 13955 98 + 1 GCCAAGGGUGGUUUUUCCUUUC----GGGAAAAACUCCCGAG--UUGCGACUACAAAACAA-----------AAUAUUCGUAGUGCCAUCGAGUACGCAAUGUUACAACCGCUCG .....((((((((.......((----((((.....))))))(--((((((((((.......-----------.......)))))((......)).)))))).....)))))))). ( -29.54) >DroEre_CAF1 7662 94 + 1 GCCAAGGGUGGUUUUUCCUUUC----GGGAAAAACUGCCGAG--UUGCGACUGAAA----A-----------AAUAUUCGUAGUGCCAUAGAGUACGCAAUGUUACAACCGCUCG ((....((..((((((((....----.))))))))..))(.(--(((..((.(((.----.-----------....)))((.((((......))))))...))..)))))))... ( -27.90) >DroYak_CAF1 30686 94 + 1 GCCAAGGGUGGUUUUUCCUUUC----GGGAAAAACUCCCGAG--CUGCGACUGAAA----A-----------AGUAUUCGUAGUGCCAUAGAGUACGCAAUGUUACAACCGCUCG .....(((.(((((((((....----.))))))))))))(((--(((..((.....----.-----------.((((((...........)))))).....))..))...)))). ( -29.00) >DroAna_CAF1 6345 108 + 1 UCCGAGGGUGGUUUUUCAUUUUCCGAAGGAAAAACUCCCUCGGAAAAUGACAAAAAAAAAA---AAAAUA---GAAAUCGUAGUGU-AUAGAGUACGCAAUAUUACAACCACUCG .....((((((((..(((((((((((.(((.....))).)))))))))))...........---......---.........((((-((...))))))........)))))))). ( -34.50) >consensus GCCAAGGGUGGUUUUUCCUUUC____GGGAAAAACUCCCGAG__UUGCGACUAAAAAA_AA___________AAUAUUCGUAGUGCCAUAGAGUACGCAAUGUUACAACCGCUCG .....((((((((((((((.......)))))).((((.......((((((...........................)))))).......))))............)))))))). (-18.09 = -18.29 + 0.20)

| Location | 16,006,008 – 16,006,112 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.46 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -21.26 |
| Energy contribution | -22.26 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16006008 104 + 23771897 UUUUUCCAGUGCGCAGAAUCGCAGUGAGUAUUAGUAUCAAAUCCUAAUCCUAUCCAAAACAGCACCACCA-------AAGGCAAAGCUAAGUGGGUGGUGGUGUUUCCUUU ......((.((((......)))).))((.(((((.........))))).)).....(((((.(((((((.-------..(((...))).....))))))).)))))..... ( -27.60) >DroSec_CAF1 6313 96 + 1 UUUUCCCAGUGCGCCGAAUCGCAGUGAGUAUUAGUAU-----CCUAAUCG----CAAAACAGCACCACCGG------UAGGCAAAGCUAAGUGGGUGGUGCUGUUUUCUUU ......((.((((......)))).)).(((((((...-----.))))).)----)((((((((((((((.(------..(((...)))...).)))))))))))))).... ( -34.00) >DroSim_CAF1 14053 96 + 1 UUUUCCCAGUGCGCCGAAUCGCAGUGAGUAUUAGUAU-----CCUAAUCG----CAAAACAGCACCACCGG------AAGGCAAAGCUAAGUGGGUGGUGCUGUUUUCUUU ......((.((((......)))).)).(((((((...-----.))))).)----)((((((((((((((.(------..(((...)))...).)))))))))))))).... ( -35.10) >DroEre_CAF1 7756 93 + 1 UUUUCGCAGUGCGCCGAAUCGCAGUGAGUAUUAGUAU-----UCUAUUCU----CAAAACAGCACAGCUUUUGGUGGUAAGCAGUAGUGAGUGGGUGGUGUU--------- ..(((((.((((((......))..((((...(((...-----.)))..))----)).....)))).((((........))))....)))))...........--------- ( -20.60) >consensus UUUUCCCAGUGCGCCGAAUCGCAGUGAGUAUUAGUAU_____CCUAAUCG____CAAAACAGCACCACCGG______AAGGCAAAGCUAAGUGGGUGGUGCUGUUUUCUUU ......((.((((......)))).))..............................(((((((((((((........................)))))))))))))..... (-21.26 = -22.26 + 1.00)

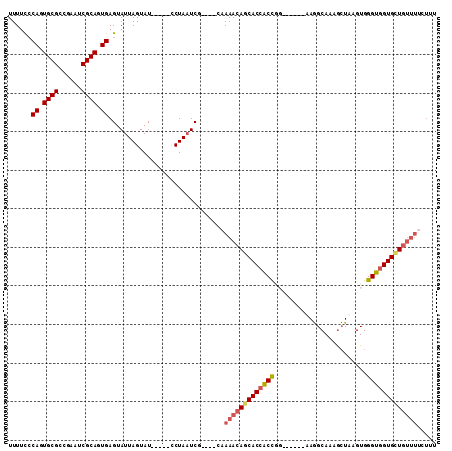
| Location | 16,006,008 – 16,006,112 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.46 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -19.61 |
| Energy contribution | -22.73 |
| Covariance contribution | 3.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16006008 104 - 23771897 AAAGGAAACACCACCACCCACUUAGCUUUGCCUU-------UGGUGGUGCUGUUUUGGAUAGGAUUAGGAUUUGAUACUAAUACUCACUGCGAUUCUGCGCACUGGAAAAA ....((((((.(((((((......((...))...-------.))))))).)))))).......(((((.........)))))..(((.((((......)))).)))..... ( -24.40) >DroSec_CAF1 6313 96 - 1 AAAGAAAACAGCACCACCCACUUAGCUUUGCCUA------CCGGUGGUGCUGUUUUG----CGAUUAGG-----AUACUAAUACUCACUGCGAUUCGGCGCACUGGGAAAA ....((((((((((((((....(((......)))------..)))))))))))))).----..(((((.-----...))))).((((.((((......)))).)))).... ( -35.50) >DroSim_CAF1 14053 96 - 1 AAAGAAAACAGCACCACCCACUUAGCUUUGCCUU------CCGGUGGUGCUGUUUUG----CGAUUAGG-----AUACUAAUACUCACUGCGAUUCGGCGCACUGGGAAAA ....((((((((((((((......((...))...------..)))))))))))))).----..(((((.-----...))))).((((.((((......)))).)))).... ( -35.10) >DroEre_CAF1 7756 93 - 1 ---------AACACCACCCACUCACUACUGCUUACCACCAAAAGCUGUGCUGUUUUG----AGAAUAGA-----AUACUAAUACUCACUGCGAUUCGGCGCACUGCGAAAA ---------..................................((.((((.....((----((..(((.-----...)))...))))..((......)))))).))..... ( -13.50) >consensus AAAGAAAACAGCACCACCCACUUAGCUUUGCCUA______CCGGUGGUGCUGUUUUG____CGAUUAGG_____AUACUAAUACUCACUGCGAUUCGGCGCACUGGGAAAA ....((((((((((((((........................)))))))))))))).......(((((.........))))).((((.((((......)))).)))).... (-19.61 = -22.73 + 3.12)


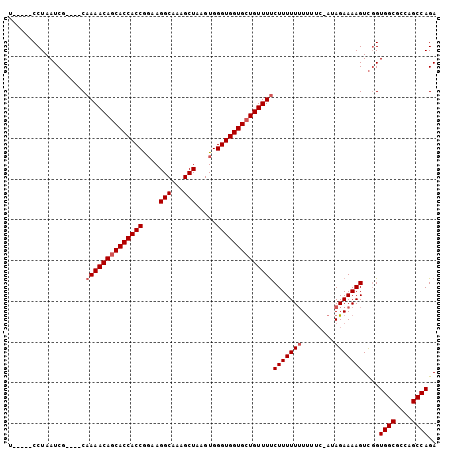
| Location | 16,006,044 – 16,006,144 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -29.00 |
| Energy contribution | -30.00 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16006044 100 + 23771897 UCAAAUCCUAAUCCUAUCCAAAACAGCACCACCA-AAGGCAAAGCUAAGUGGGUGGUGGUGUUUCCUUUUUUUCU----UAGAAAAGUUCGUGGCGCCAGCCAGA ....................(((((.(((((((.-..(((...))).....))))))).))))).(((((((...----.)))))))....((((....)))).. ( -27.80) >DroSec_CAF1 6349 95 + 1 U-----CCUAAUCG----CAAAACAGCACCACCGGUAGGCAAAGCUAAGUGGGUGGUGCUGUUUUCUUUUU-UUUUCCAUAGAAAAGUCGGUGGCGCCAGCCAGA .-----((......----.((((((((((((((.(..(((...)))...).))))))))))))))((((((-(.......)))))))..))((((....)))).. ( -33.80) >DroSim_CAF1 14089 96 + 1 U-----CCUAAUCG----CAAAACAGCACCACCGGAAGGCAAAGCUAAGUGGGUGGUGCUGUUUUCUUUUUUUUUUCUAUAGAAAAGUCGGUGGCGCCAGCCAGA .-----....((((----(((((((((((((((.(..(((...)))...).)))))))))))))).......(((((....)))))).))))(((....)))... ( -35.20) >consensus U_____CCUAAUCG____CAAAACAGCACCACCGGAAGGCAAAGCUAAGUGGGUGGUGCUGUUUUCUUUUUUUUUUC_AUAGAAAAGUCGGUGGCGCCAGCCAGA ...................((((((((((((((....(((...))).....))))))))))))))(((((((........)))))))....((((....)))).. (-29.00 = -30.00 + 1.00)


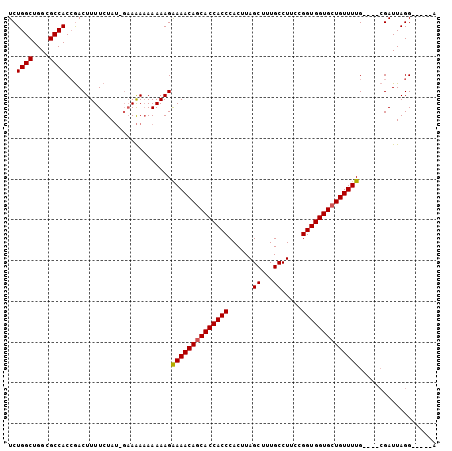
| Location | 16,006,044 – 16,006,144 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -25.79 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 16006044 100 - 23771897 UCUGGCUGGCGCCACGAACUUUUCUA----AGAAAAAAAGGAAACACCACCACCCACUUAGCUUUGCCUU-UGGUGGUGCUGUUUUGGAUAGGAUUAGGAUUUGA ((((((....)))).))....(((((----(.........((((((.(((((((......((...))...-.))))))).))))))........))))))..... ( -24.83) >DroSec_CAF1 6349 95 - 1 UCUGGCUGGCGCCACCGACUUUUCUAUGGAAAA-AAAAAGAAAACAGCACCACCCACUUAGCUUUGCCUACCGGUGGUGCUGUUUUG----CGAUUAGG-----A ..((((....))))((...(((((....)))))-......((((((((((((((....(((......)))..)))))))))))))).----......))-----. ( -31.90) >DroSim_CAF1 14089 96 - 1 UCUGGCUGGCGCCACCGACUUUUCUAUAGAAAAAAAAAAGAAAACAGCACCACCCACUUAGCUUUGCCUUCCGGUGGUGCUGUUUUG----CGAUUAGG-----A ..((((....))))((...(((((....))))).......((((((((((((((......((...)).....)))))))))))))).----......))-----. ( -31.40) >consensus UCUGGCUGGCGCCACCGACUUUUCUAU_GAAAAAAAAAAGAAAACAGCACCACCCACUUAGCUUUGCCUUCCGGUGGUGCUGUUUUG____CGAUUAGG_____A ..((((....))))..........................((((((((((((((......((...)).....))))))))))))))................... (-25.79 = -25.90 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:14 2006