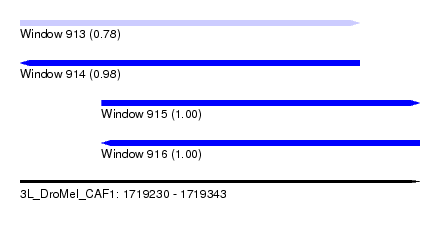
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,719,230 – 1,719,343 |
| Length | 113 |
| Max. P | 0.999868 |

| Location | 1,719,230 – 1,719,326 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 85.37 |
| Mean single sequence MFE | -37.44 |
| Consensus MFE | -31.44 |
| Energy contribution | -30.84 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
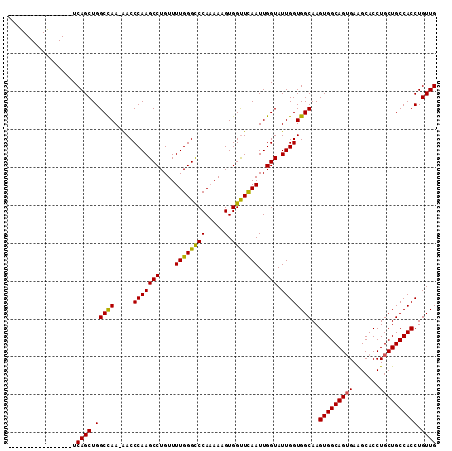
>3L_DroMel_CAF1 1719230 96 + 23771897 -----------------UCAGCUGGCCAAAGACCCAAGCCUGUUUUGGGCCCAAAAAGUGGUUCAAUUGGUAUUGGUGGCAAGUGGCAGUGAAGCACCUGCUGCCACCUGUUG -----------------.((((.(((((.....(((((((....((((((((.....).)))))))..))).))))))))..((((((((.........))))))))).)))) ( -33.70) >DroSec_CAF1 1155 95 + 1 -----------------UCAGCUGGCCAA-AACCCAAGCCUGUUUUGGGCCCAAAAGGUGGUUCAAUUGGUAUUGGUGGCAAGUGGCAGUGAGGCACCUGCUGCCACCUGUUG -----------------.((((.(((((.-...(((((((....((((((((.....).)))))))..))).))))))))..((((((((.((....))))))))))).)))) ( -36.30) >DroSim_CAF1 1124 95 + 1 -----------------UCAGCUGGCCAA-GACCCAAGCCUGUUUUGGGUCCAAAAGGUGGUUCAAUUGGUAUUGGUGGCAAGUGGCAGUGAGGCACCUGCUGCCACCUGUUG -----------------.(((.((((...-((((((((.....))))))))....(((((.((((.(((.((((.......)))).))))))).)))))...)))).)))... ( -35.30) >DroEre_CAF1 1180 112 + 1 UCAUGGCCACGCAGCGCUCAGCUGGCUAA-AACCCAAGCCUGUUUUGGGCCCAUAAAGUGGCUUAAUUGGUAUUGGUGGCAAGUGGCAGGGAAGCACCUGCUGCCACCUGUUG (((.((((((.((((.....)))).....-..((((((.....))))))........))))))....)))....(((((((....(((((......))))))))))))..... ( -43.60) >DroYak_CAF1 1032 112 + 1 UUACGGCCACGCAGCUCUCAGCUGGCUAA-AACCCAAGCCUGUUUUGGGCCCAUAAAGUGGUUUAAUUGGUAUUGGUGGCAAGUGGCAGUGAAGCACCUGCUGCCACCUGUUG .....(((((.((((.....))))(((((-..((((((.....)))))).((((...)))).....)))))....)))))..((((((((.........))))))))...... ( -38.30) >consensus _________________UCAGCUGGCCAA_AACCCAAGCCUGUUUUGGGCCCAAAAAGUGGUUCAAUUGGUAUUGGUGGCAAGUGGCAGUGAAGCACCUGCUGCCACCUGUUG ..................((((.(((((.....(((((((....((((((((.....).)))))))..))).))))))))..(((((((((.......)))))))))).)))) (-31.44 = -30.84 + -0.60)

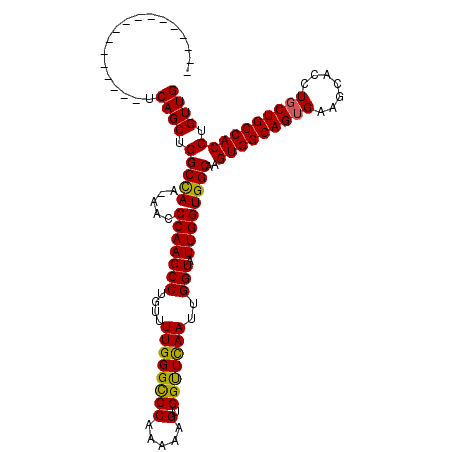
| Location | 1,719,230 – 1,719,326 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.37 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -30.30 |
| Energy contribution | -29.90 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1719230 96 - 23771897 CAACAGGUGGCAGCAGGUGCUUCACUGCCACUUGCCACCAAUACCAAUUGAACCACUUUUUGGGCCCAAAACAGGCUUGGGUCUUUGGCCAGCUGA----------------- ...(((.((((.((((((((......).)))))))..........................((((((((.......))))))))...)))).))).----------------- ( -32.50) >DroSec_CAF1 1155 95 - 1 CAACAGGUGGCAGCAGGUGCCUCACUGCCACUUGCCACCAAUACCAAUUGAACCACCUUUUGGGCCCAAAACAGGCUUGGGUU-UUGGCCAGCUGA----------------- ...(((.((((.(((((((.(.....).)))))))...(((..((((..(......)..))))((((((.......)))))).-))))))).))).----------------- ( -33.30) >DroSim_CAF1 1124 95 - 1 CAACAGGUGGCAGCAGGUGCCUCACUGCCACUUGCCACCAAUACCAAUUGAACCACCUUUUGGACCCAAAACAGGCUUGGGUC-UUGGCCAGCUGA----------------- ...(((.((((.(((((((.(.....).)))))))...(((...(((..(......)..)))(((((((.......)))))))-))))))).))).----------------- ( -33.00) >DroEre_CAF1 1180 112 - 1 CAACAGGUGGCAGCAGGUGCUUCCCUGCCACUUGCCACCAAUACCAAUUAAGCCACUUUAUGGGCCCAAAACAGGCUUGGGUU-UUAGCCAGCUGAGCGCUGCGUGGCCAUGA .....((((((((((((......)))))....)))))))............(((((.....((((((((.......)))))))-)..((.(((.....))))))))))..... ( -42.00) >DroYak_CAF1 1032 112 - 1 CAACAGGUGGCAGCAGGUGCUUCACUGCCACUUGCCACCAAUACCAAUUAAACCACUUUAUGGGCCCAAAACAGGCUUGGGUU-UUAGCCAGCUGAGAGCUGCGUGGCCGUAA ..((.(((((((((((........))))....))))))).............((((.....((((((((.......)))))))-)..((.(((.....)))))))))..)).. ( -36.80) >consensus CAACAGGUGGCAGCAGGUGCUUCACUGCCACUUGCCACCAAUACCAAUUGAACCACUUUUUGGGCCCAAAACAGGCUUGGGUU_UUGGCCAGCUGA_________________ ...(((.((((.((((((((......).)))))))...........................(((((((.......)))))))....)))).))).................. (-30.30 = -29.90 + -0.40)

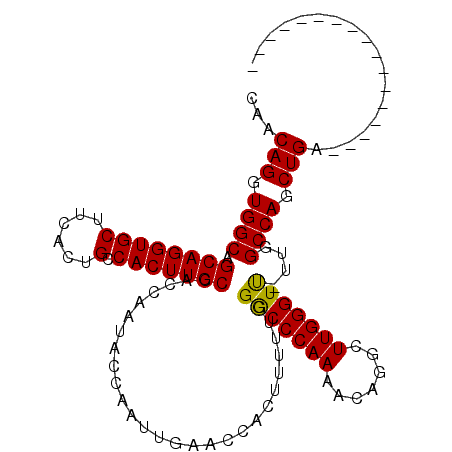
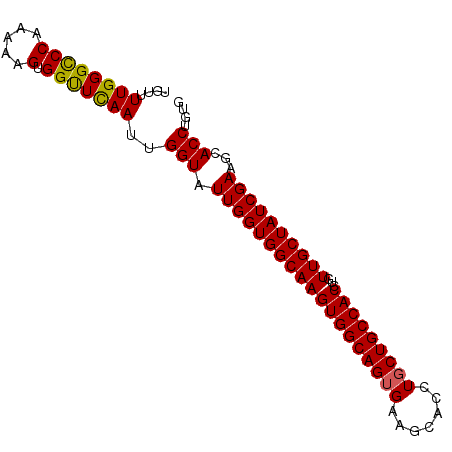
| Location | 1,719,253 – 1,719,343 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -30.92 |
| Energy contribution | -30.56 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1719253 90 + 23771897 UGUUUUGGGCCCAAAAAGUGGUUCAAUUGGUAUUGGUGGCAAGUGGCAGUGAAGCACCUGCUGCCACCUGUUGCUAUCGAAGCACCUGUG ....((((((((.....).)))))))..(((.((((((((((((((((((.........))))))))...))))))))))...))).... ( -32.50) >DroSec_CAF1 1177 90 + 1 UGUUUUGGGCCCAAAAGGUGGUUCAAUUGGUAUUGGUGGCAAGUGGCAGUGAGGCACCUGCUGCCACCUGUUGCUAUCGAUGCACCUGUG ....((((((((.....).)))))))...(((((((((((((((((((((.((....))))))))))...)))))))))))))....... ( -38.40) >DroSim_CAF1 1146 90 + 1 UGUUUUGGGUCCAAAAGGUGGUUCAAUUGGUAUUGGUGGCAAGUGGCAGUGAGGCACCUGCUGCCACCUGUUGCUAUCGAUGCACCUGUG ....(((((.(((.....))))))))...(((((((((((((((((((((.((....))))))))))...)))))))))))))....... ( -36.10) >DroEre_CAF1 1219 90 + 1 UGUUUUGGGCCCAUAAAGUGGCUUAAUUGGUAUUGGUGGCAAGUGGCAGGGAAGCACCUGCUGCCACCUGUUGCUAUCGAAGCACCUGUG ((((((((((((.....).))))....(((((..(((((((....(((((......))))))))))))...))))).)))))))...... ( -35.30) >DroYak_CAF1 1071 90 + 1 UGUUUUGGGCCCAUAAAGUGGUUUAAUUGGUAUUGGUGGCAAGUGGCAGUGAAGCACCUGCUGCCACCUGUUGCUAUCGAAGUACCUGUG ....((((((((.....).)))))))..((((((((((((((((((((((.........))))))))...)))))))))..))))).... ( -32.10) >consensus UGUUUUGGGCCCAAAAAGUGGUUCAAUUGGUAUUGGUGGCAAGUGGCAGUGAAGCACCUGCUGCCACCUGUUGCUAUCGAAGCACCUGUG ....((((((((.....).)))))))..(((.(((((((((((((((((((.......)))))))))...))))))))))...))).... (-30.92 = -30.56 + -0.36)

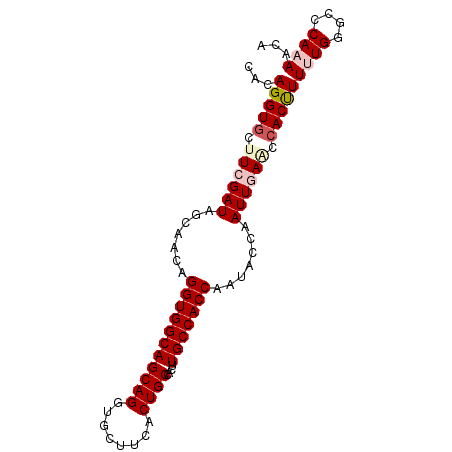
| Location | 1,719,253 – 1,719,343 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -24.64 |
| Energy contribution | -25.72 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1719253 90 - 23771897 CACAGGUGCUUCGAUAGCAACAGGUGGCAGCAGGUGCUUCACUGCCACUUGCCACCAAUACCAAUUGAACCACUUUUUGGGCCCAAAACA ...(((((.((((((.......(((((((((((........))))....))))))).......)))))).)))))((((....))))... ( -27.74) >DroSec_CAF1 1177 90 - 1 CACAGGUGCAUCGAUAGCAACAGGUGGCAGCAGGUGCCUCACUGCCACUUGCCACCAAUACCAAUUGAACCACCUUUUGGGCCCAAAACA ...(((((..(((((.......(((((((((((........))))....))))))).......)))))..)))))((((....))))... ( -28.14) >DroSim_CAF1 1146 90 - 1 CACAGGUGCAUCGAUAGCAACAGGUGGCAGCAGGUGCCUCACUGCCACUUGCCACCAAUACCAAUUGAACCACCUUUUGGACCCAAAACA ...(((((..(((((.......(((((((((((........))))....))))))).......)))))..)))))((((....))))... ( -28.14) >DroEre_CAF1 1219 90 - 1 CACAGGUGCUUCGAUAGCAACAGGUGGCAGCAGGUGCUUCCCUGCCACUUGCCACCAAUACCAAUUAAGCCACUUUAUGGGCCCAAAACA ....((((((.....)))....((((((((((((......)))))....)))))))...)))......(((.(.....))))........ ( -29.90) >DroYak_CAF1 1071 90 - 1 CACAGGUACUUCGAUAGCAACAGGUGGCAGCAGGUGCUUCACUGCCACUUGCCACCAAUACCAAUUAAACCACUUUAUGGGCCCAAAACA ....(((...............(((((((((((........))))....))))))).............(((.....))))))....... ( -23.10) >consensus CACAGGUGCUUCGAUAGCAACAGGUGGCAGCAGGUGCUUCACUGCCACUUGCCACCAAUACCAAUUGAACCACUUUUUGGGCCCAAAACA ...(((((.((((((.......(((((((((((........))))....))))))).......)))))).)))))((((....))))... (-24.64 = -25.72 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:18 2006