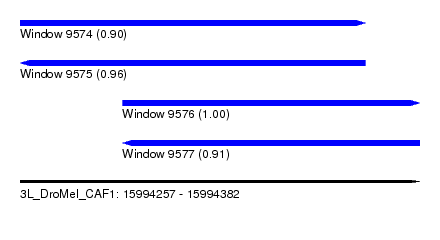
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,994,257 – 15,994,382 |
| Length | 125 |
| Max. P | 0.999439 |

| Location | 15,994,257 – 15,994,365 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.24 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -13.96 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15994257 108 + 23771897 AAGGGAAAGGU----GGGGGAAAA---CAACACAACGAA----ACGGCAGGCGGCAAAAAUUUCUUUGAUCUUAUCGCAAAGCUUCUUGCUCGCAUUUUGAUCAAAGCAA-GUUUUCCGC .........((----(..(.....---)..)))......----.......((((..(((((((((((((((..(..((..(((.....))).))..)..)))))))).))-))))))))) ( -26.90) >DroVir_CAF1 4876 90 + 1 -------------------AACAA---AAACGUAACGAA----AU---GCGCGGCAAAAAUAUCUUUGAUCUUAUCGCAAAGCUUCUUGCUCGCAUUUUGAUCAAAGCAA-UUUUUCCGC -------------------.....---....(((.....----.)---))((((.((((((..((((((((..(..((..(((.....))).))..)..))))))))..)-))))))))) ( -23.10) >DroPse_CAF1 14575 100 + 1 AAGGCAAAGGA----AC--GAAAGCGCCAGCACAACGAUUAAAACUUUUUGCGGUGGC----UCUUUCAUUCCUGCCCCGUGCCCCGUGCCC-CAU----GCCAAAGC-----CUUCUCC ..((((.((((----(.--(((((.((((.(.(((.((.......)).))).).))))----.))))).))))).....(..(...)..)..-..)----))).....-----....... ( -28.60) >DroSim_CAF1 2238 108 + 1 AAGGGAAAGGU----GGGGGAAAA---CAACACAACGAA----ACGGCAGGCGGCAAAAAUUUCUUUGAUCUUAUCGCAAAGCUUCUUGCUCGCAUUUUGAUCAAAGCAA-GUUUUCCGC .........((----(..(.....---)..)))......----.......((((..(((((((((((((((..(..((..(((.....))).))..)..)))))))).))-))))))))) ( -26.90) >DroYak_CAF1 17907 112 + 1 AAGGAAAAGGGGGGAGGGGGAAAA---UAACACAACGAA----ACGGCAGGCGGCAACAAUUUCUUUGAUCUUAUCGCAAAGCUUCUUGCUCGCAUUUUGAUCAAAGCAA-GUUUUCCGC ..................((((((---(...........----.........(....).....((((((((..(..((..(((.....))).))..)..))))))))...-))))))).. ( -21.70) >DroMoj_CAF1 4529 91 + 1 -------------------AACAA---AAACGUAACGAA----AU---ACGCGGCAAAAAUUUCUUUGAUCUUAUCGCAAAGCUUCUUGCUCGCAUUUUGAUCAAAGCAAGUUUUUCCGC -------------------.....---....(((.....----.)---))((((.((((((((((((((((..(..((..(((.....))).))..)..)))))))).)))))))))))) ( -26.00) >consensus AAGG_AAAGG_____GG__GAAAA___CAACACAACGAA____ACGGCAGGCGGCAAAAAUUUCUUUGAUCUUAUCGCAAAGCUUCUUGCUCGCAUUUUGAUCAAAGCAA_GUUUUCCGC ..................................................((((.(((.....((((((((.....((..(((.....))).)).....)))))))).....))).)))) (-13.96 = -14.77 + 0.81)


| Location | 15,994,257 – 15,994,365 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.24 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -18.25 |
| Energy contribution | -19.17 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15994257 108 - 23771897 GCGGAAAAC-UUGCUUUGAUCAAAAUGCGAGCAAGAAGCUUUGCGAUAAGAUCAAAGAAAUUUUUGCCGCCUGCCGU----UUCGUUGUGUUG---UUUUCCCCC----ACCUUUCCCUU ..(((((((-.(((..(((.......(((.(((((((.(((((.........)))))...)))))))))).......----.)))..)))..)---))))))...----........... ( -25.56) >DroVir_CAF1 4876 90 - 1 GCGGAAAAA-UUGCUUUGAUCAAAAUGCGAGCAAGAAGCUUUGCGAUAAGAUCAAAGAUAUUUUUGCCGCGC---AU----UUCGUUACGUUU---UUGUU------------------- (((((((((-(..((((((((....(((((((.....)).)))))....))))))))..)))))).))))..---..----............---.....------------------- ( -26.40) >DroPse_CAF1 14575 100 - 1 GGAGAAG-----GCUUUGGC----AUG-GGGCACGGGGCACGGGGCAGGAAUGAAAGA----GCCACCGCAAAAAGUUUUAAUCGUUGUGCUGGCGCUUUC--GU----UCCUUUGCCUU .......-----((((((((----...-..)).))))))..((((((((((((((((.----((((.(((((...(.......).))))).)))).)))))--))----)))..)))))) ( -41.00) >DroSim_CAF1 2238 108 - 1 GCGGAAAAC-UUGCUUUGAUCAAAAUGCGAGCAAGAAGCUUUGCGAUAAGAUCAAAGAAAUUUUUGCCGCCUGCCGU----UUCGUUGUGUUG---UUUUCCCCC----ACCUUUCCCUU ..(((((((-.(((..(((.......(((.(((((((.(((((.........)))))...)))))))))).......----.)))..)))..)---))))))...----........... ( -25.56) >DroYak_CAF1 17907 112 - 1 GCGGAAAAC-UUGCUUUGAUCAAAAUGCGAGCAAGAAGCUUUGCGAUAAGAUCAAAGAAAUUGUUGCCGCCUGCCGU----UUCGUUGUGUUA---UUUUCCCCCUCCCCCCUUUUCCUU ((((..(((-...((((((((....(((((((.....)).)))))....)))))))).....))).)))).......----............---........................ ( -23.00) >DroMoj_CAF1 4529 91 - 1 GCGGAAAAACUUGCUUUGAUCAAAAUGCGAGCAAGAAGCUUUGCGAUAAGAUCAAAGAAAUUUUUGCCGCGU---AU----UUCGUUACGUUU---UUGUU------------------- (((((((((.((.((((((((....(((((((.....)).)))))....)))))))))).))))).))))((---(.----.....)))....---.....------------------- ( -26.00) >consensus GCGGAAAAC_UUGCUUUGAUCAAAAUGCGAGCAAGAAGCUUUGCGAUAAGAUCAAAGAAAUUUUUGCCGCCUGCCGU____UUCGUUGUGUUG___UUUUC__CC_____CCUUU_CCUU ((((.........((((((((....(((((((.....)).)))))....)))))))).........)))).................................................. (-18.25 = -19.17 + 0.92)


| Location | 15,994,289 – 15,994,382 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.40 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -21.92 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.56 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15994289 93 + 23771897 ACGGCAGGCGGCAAAAAUUUCUUUGAUCUUAUCGCAAAGCUUCUUGCUCGCAUUUUGAUCAAAGCAA-GUUUUCCGCCAAAAACACACACACAC------------ ......(((((..(((((((((((((((..(..((..(((.....))).))..)..)))))))).))-))))))))))................------------ ( -27.70) >DroVir_CAF1 4893 97 + 1 AU---GCGCGGCAAAAAUAUCUUUGAUCUUAUCGCAAAGCUUCUUGCUCGCAUUUUGAUCAAAGCAA-UUUUUCCGCCAAAA-UACAAAGGCAACAACAACA---- .(---(.((((.((((((..((((((((..(..((..(((.....))).))..)..))))))))..)-)))))))))))...-......(....).......---- ( -25.80) >DroSim_CAF1 2270 89 + 1 ACGGCAGGCGGCAAAAAUUUCUUUGAUCUUAUCGCAAAGCUUCUUGCUCGCAUUUUGAUCAAAGCAA-GUUUUCCGCCAAAAACACACAC---------------- ......(((((..(((((((((((((((..(..((..(((.....))).))..)..)))))))).))-))))))))))............---------------- ( -27.70) >DroYak_CAF1 17943 91 + 1 ACGGCAGGCGGCAACAAUUUCUUUGAUCUUAUCGCAAAGCUUCUUGCUCGCAUUUUGAUCAAAGCAA-GUUUUCCGCCAAAAACACACACAC-------------- ......(((((.((.(((((((((((((..(..((..(((.....))).))..)..)))))))).))-))))))))))..............-------------- ( -25.80) >DroMoj_CAF1 4546 102 + 1 AU---ACGCGGCAAAAAUUUCUUUGAUCUUAUCGCAAAGCUUCUUGCUCGCAUUUUGAUCAAAGCAAGUUUUUCCGCCAAAA-UACAAAGGCAACAACAACAACAU ..---..((((.((((((((((((((((..(..((..(((.....))).))..)..)))))))).)))))))))))).....-......(....)........... ( -27.60) >consensus ACGGCAGGCGGCAAAAAUUUCUUUGAUCUUAUCGCAAAGCUUCUUGCUCGCAUUUUGAUCAAAGCAA_GUUUUCCGCCAAAAACACACACACA_____________ ......(((((.((((....((((((((..(..((..(((.....))).))..)..)))))))).....)))))))))............................ (-21.92 = -22.52 + 0.60)



| Location | 15,994,289 – 15,994,382 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.40 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -24.27 |
| Energy contribution | -24.67 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15994289 93 - 23771897 ------------GUGUGUGUGUGUUUUUGGCGGAAAAC-UUGCUUUGAUCAAAAUGCGAGCAAGAAGCUUUGCGAUAAGAUCAAAGAAAUUUUUGCCGCCUGCCGU ------------................(((((((((.-((.((((((((....(((((((.....)).)))))....)))))))).)).)))).)))))...... ( -27.00) >DroVir_CAF1 4893 97 - 1 ----UGUUGUUGUUGCCUUUGUA-UUUUGGCGGAAAAA-UUGCUUUGAUCAAAAUGCGAGCAAGAAGCUUUGCGAUAAGAUCAAAGAUAUUUUUGCCGCGC---AU ----...................-...(((((((((((-(..((((((((....(((((((.....)).)))))....))))))))..)))))).)))).)---). ( -26.60) >DroSim_CAF1 2270 89 - 1 ----------------GUGUGUGUUUUUGGCGGAAAAC-UUGCUUUGAUCAAAAUGCGAGCAAGAAGCUUUGCGAUAAGAUCAAAGAAAUUUUUGCCGCCUGCCGU ----------------............(((((((((.-((.((((((((....(((((((.....)).)))))....)))))))).)).)))).)))))...... ( -27.00) >DroYak_CAF1 17943 91 - 1 --------------GUGUGUGUGUUUUUGGCGGAAAAC-UUGCUUUGAUCAAAAUGCGAGCAAGAAGCUUUGCGAUAAGAUCAAAGAAAUUGUUGCCGCCUGCCGU --------------..............(((((..(((-...((((((((....(((((((.....)).)))))....)))))))).....))).)))))...... ( -26.70) >DroMoj_CAF1 4546 102 - 1 AUGUUGUUGUUGUUGCCUUUGUA-UUUUGGCGGAAAAACUUGCUUUGAUCAAAAUGCGAGCAAGAAGCUUUGCGAUAAGAUCAAAGAAAUUUUUGCCGCGU---AU .......................-.....(((((((((.((.((((((((....(((((((.....)).)))))....)))))))))).))))).))))..---.. ( -24.80) >consensus _____________UGCGUGUGUGUUUUUGGCGGAAAAC_UUGCUUUGAUCAAAAUGCGAGCAAGAAGCUUUGCGAUAAGAUCAAAGAAAUUUUUGCCGCCUGCCGU ............................(((((.........((((((((....(((((((.....)).)))))....)))))))).........)))))...... (-24.27 = -24.67 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:07 2006