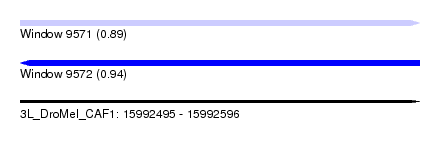
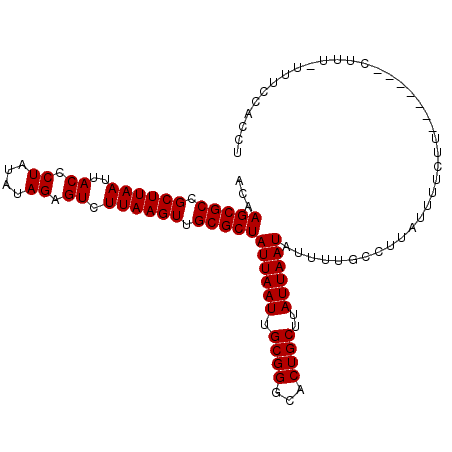
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,992,495 – 15,992,596 |
| Length | 101 |
| Max. P | 0.941739 |

| Location | 15,992,495 – 15,992,596 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.38 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15992495 101 + 23771897 AGGUGGAAA-AAAG-------AAGAAAAUAAGGCAAAAUAUUAAUAAGCAGUGCCCGCAAUUAAUAGCGCAACUUAAGACUCUAUAUAGGGUAAUUAAGCGGCGCUUGU .........-....-------...........((((..(((((((..((.......)).)))))))((((..(((((.(((((....)))))..)))))..)))))))) ( -20.30) >DroVir_CAF1 2854 106 + 1 AAGCAGAAA-AGAGUCGCUACAAGCC--AAAAGCAAAAUAUUAAUAAGCAGUGCCCGCAAUUAAUAGCGCAACUUAAGACGCUAUAUAGGGUAAUUAAGCGGCGCUUGU ........(-((.((((((....((.--....)).................(((((.......(((((((.......).))))))...)))))....)))))).))).. ( -24.10) >DroSim_CAF1 465 102 + 1 AGGUGGAAAAAAAG-------AAGAAAAUAAGGCAAAAUAUUAAUAAGCAGUGCCCGCAAUUAAUAGCGCAACUUAAGACUCUAUAUAGGGUAAUUAAGCGGCGCUUGU ..............-------...........((((..(((((((..((.......)).)))))))((((..(((((.(((((....)))))..)))))..)))))))) ( -20.30) >DroYak_CAF1 16053 101 + 1 AGGUGGAAA-AAAG-------AAGAAAAUAAGGCAAAAUAUUAAUAAGCAGUGCCCGCAAUUAAUAGCGCAACUUAAGACUCUAUAUAGGGUAAUUAAGCGGCGCUUGU .........-....-------...........((((..(((((((..((.......)).)))))))((((..(((((.(((((....)))))..)))))..)))))))) ( -20.30) >consensus AGGUGGAAA_AAAG_______AAGAAAAUAAGGCAAAAUAUUAAUAAGCAGUGCCCGCAAUUAAUAGCGCAACUUAAGACUCUAUAUAGGGUAAUUAAGCGGCGCUUGU ................................((((..(((((((..((.......)).)))))))((((..(((((.(((((....)))))..)))))..)))))))) (-19.12 = -19.38 + 0.25)

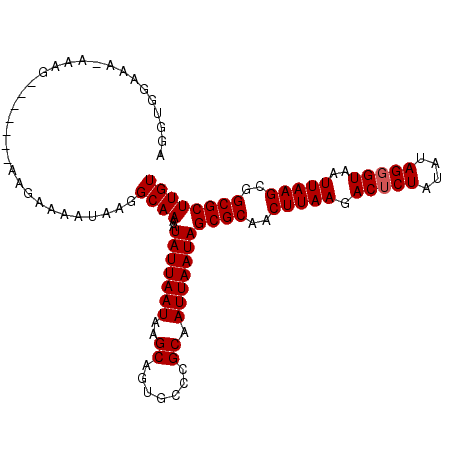

| Location | 15,992,495 – 15,992,596 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -19.63 |
| Consensus MFE | -18.45 |
| Energy contribution | -18.45 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15992495 101 - 23771897 ACAAGCGCCGCUUAAUUACCCUAUAUAGAGUCUUAAGUUGCGCUAUUAAUUGCGGGCACUGCUUAUUAAUAUUUUGCCUUAUUUUCUU-------CUUU-UUUCCACCU ...(((((.((((((..((.((....)).)).)))))).)))))((((((.((((...))))..))))))..................-------....-......... ( -18.50) >DroVir_CAF1 2854 106 - 1 ACAAGCGCCGCUUAAUUACCCUAUAUAGCGUCUUAAGUUGCGCUAUUAAUUGCGGGCACUGCUUAUUAAUAUUUUGCUUUU--GGCUUGUAGCGACUCU-UUUCUGCUU (((((((((.(.((((((......(((((((........))))))))))))).))))...((.............))....--.))))))(((((....-..)).))). ( -23.02) >DroSim_CAF1 465 102 - 1 ACAAGCGCCGCUUAAUUACCCUAUAUAGAGUCUUAAGUUGCGCUAUUAAUUGCGGGCACUGCUUAUUAAUAUUUUGCCUUAUUUUCUU-------CUUUUUUUCCACCU ...(((((.((((((..((.((....)).)).)))))).)))))((((((.((((...))))..))))))..................-------.............. ( -18.50) >DroYak_CAF1 16053 101 - 1 ACAAGCGCCGCUUAAUUACCCUAUAUAGAGUCUUAAGUUGCGCUAUUAAUUGCGGGCACUGCUUAUUAAUAUUUUGCCUUAUUUUCUU-------CUUU-UUUCCACCU ...(((((.((((((..((.((....)).)).)))))).)))))((((((.((((...))))..))))))..................-------....-......... ( -18.50) >consensus ACAAGCGCCGCUUAAUUACCCUAUAUAGAGUCUUAAGUUGCGCUAUUAAUUGCGGGCACUGCUUAUUAAUAUUUUGCCUUAUUUUCUU_______CUUU_UUUCCACCU ...(((((.((((((..((.((....)).)).)))))).)))))((((((.((((...))))..))))))....................................... (-18.45 = -18.45 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:03 2006