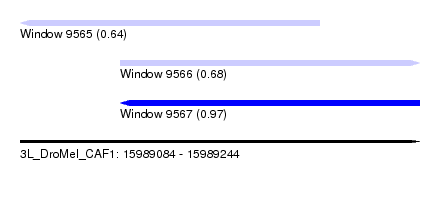
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,989,084 – 15,989,244 |
| Length | 160 |
| Max. P | 0.970111 |

| Location | 15,989,084 – 15,989,204 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -23.24 |
| Energy contribution | -23.08 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15989084 120 - 23771897 UGCGGCGUUCGCUUUUGAUAACUCACCUGCUGUGGCUUAAUCCAUUUAUUUGUAGCUUCUCCGGCUUUAGCUGCCAUUUUAUCAAUUUGUUCGACUGCCAUUUUCGAUUCGAUUACACAU ...((((.(((...(((((((.......((((..(..(((.....))).)..))))......(((.......)))...)))))))......))).))))..................... ( -23.50) >DroSec_CAF1 18096 120 - 1 UGCGGCGUUCGCUUUUGAUAACUCACCUGCUGUGGCUUAAUCCAUUUAUGUGUAGCUUCUCCGGCUUUAGCUGCCAUUUUAUCAAUUUGUUCGACUGCCAUUUUCGAUUCGAUUACACAU ...((((.(((...((((((((((((.((..((((......)))).)).))).)).......(((.......)))...)))))))......))).))))..................... ( -23.30) >DroSim_CAF1 19236 120 - 1 UGCGGCGUUCGCUUUUGAUAACUCACCUGCUGUGGCUUAAUCCAUUUUUUUGUAGCUUCUCCGGCUUUAGCUGCCAUUUUAUCAAUUUGUUCGACUGCCAUUUUCGAUUCGAUUACACAU ...((((.(((...(((((((.....((((.((((......))))......)))).......(((.......)))...)))))))......))).))))..................... ( -22.60) >DroEre_CAF1 19117 120 - 1 UUCGGCGUUCGCUUUUGAUAACUCACCUGCUGUGGAUUAAUCCAUUUAUUUGUAGCUUCUCCGGCUUUAGCUGCCAUUUUAUCAAUUUGUUCGACUGCCAUUUUCGAUUCGAUUACACAU ...((((.(((...(((((((.......((((..(((..........)))..))))......(((.......)))...)))))))......))).))))..................... ( -25.30) >DroYak_CAF1 18630 119 - 1 UUCGGCGUUCGCUUUUGAUAACUCACCUGCUGUGGCUUAAUCCACUUAUUUGUAGCUUCUCCGGCUUUAGCUGCCAUUUUAUCAAAUUGUUCGACUGCCAUUUUCGAUUCGA-UACACAU ...((((.(((..((((((((.....((((.((((......))))......)))).......(((.......)))...)))))))).....))).)))).............-....... ( -25.20) >consensus UGCGGCGUUCGCUUUUGAUAACUCACCUGCUGUGGCUUAAUCCAUUUAUUUGUAGCUUCUCCGGCUUUAGCUGCCAUUUUAUCAAUUUGUUCGACUGCCAUUUUCGAUUCGAUUACACAU ...((((.(((...(((((((.....((((.((((......))))......)))).......(((.......)))...)))))))......))).))))..................... (-23.24 = -23.08 + -0.16)

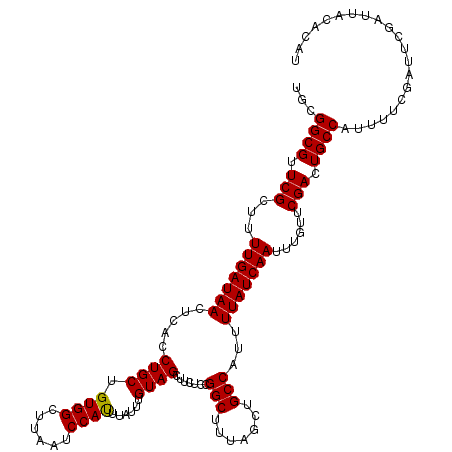

| Location | 15,989,124 – 15,989,244 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -24.84 |
| Energy contribution | -24.84 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15989124 120 + 23771897 AAAAUGGCAGCUAAAGCCGGAGAAGCUACAAAUAAAUGGAUUAAGCCACAGCAGGUGAGUUAUCAAAAGCGAACGCCGCAAUACGGUAGCCCCACAAAUGAAAUAUGAAGCUGCUGCGAA .....(((((((......((.(..(((((.......(((......)))..((.((((.(((......)))...))))))......)))))))).(....)........)))))))..... ( -26.70) >DroSec_CAF1 18136 120 + 1 AAAAUGGCAGCUAAAGCCGGAGAAGCUACACAUAAAUGGAUUAAGCCACAGCAGGUGAGUUAUCAAAAGCGAACGCCGCAAUACGGUAGCCCCACAAAUGAAAUAUGAAGCUGCUGCGAA ......(((((....((((((..((((.(((.....(((......)))......))))))).))....(((.....)))....))))(((..((...........))..))))))))... ( -27.60) >DroSim_CAF1 19276 120 + 1 AAAAUGGCAGCUAAAGCCGGAGAAGCUACAAAAAAAUGGAUUAAGCCACAGCAGGUGAGUUAUCAAAAGCGAACGCCGCAAUACGGUAGCCCCACAAAUGAAAUAUGAAGCUGCUGCGAA .....(((((((......((.(..(((((.......(((......)))..((.((((.(((......)))...))))))......)))))))).(....)........)))))))..... ( -26.70) >DroEre_CAF1 19157 120 + 1 AAAAUGGCAGCUAAAGCCGGAGAAGCUACAAAUAAAUGGAUUAAUCCACAGCAGGUGAGUUAUCAAAAGCGAACGCCGAAAUACGGUAGCCCCACAAAUGAAAUACGAAGCUGCUGCGAA .....(((((((......((.(..(((((.......((((....)))).....((((.(((......)))...))))........)))))))).(....)........)))))))..... ( -25.90) >DroYak_CAF1 18669 120 + 1 AAAAUGGCAGCUAAAGCCGGAGAAGCUACAAAUAAGUGGAUUAAGCCACAGCAGGUGAGUUAUCAAAAGCGAACGCCGAAAUACGGUAGCCCCACAAAUGAAAUAUGAAGCUGCUGCGAA .....(((((((......((.(..(((((......((((......))))....((((.(((......)))...))))........)))))))).(....)........)))))))..... ( -28.50) >consensus AAAAUGGCAGCUAAAGCCGGAGAAGCUACAAAUAAAUGGAUUAAGCCACAGCAGGUGAGUUAUCAAAAGCGAACGCCGCAAUACGGUAGCCCCACAAAUGAAAUAUGAAGCUGCUGCGAA .....(((((((......((.(..(((((.......(((......))).....((((.(((......)))...))))........)))))))).(....)........)))))))..... (-24.84 = -24.84 + -0.00)


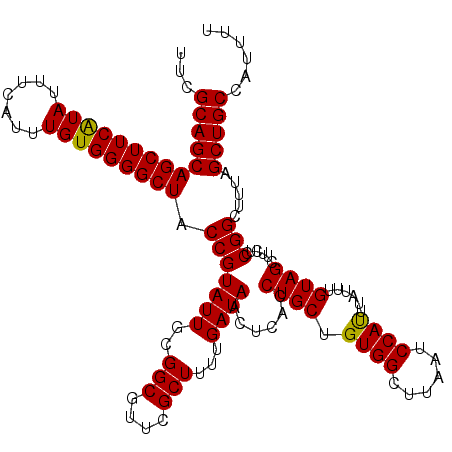
| Location | 15,989,124 – 15,989,244 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -29.24 |
| Energy contribution | -28.92 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15989124 120 - 23771897 UUCGCAGCAGCUUCAUAUUUCAUUUGUGGGGCUACCGUAUUGCGGCGUUCGCUUUUGAUAACUCACCUGCUGUGGCUUAAUCCAUUUAUUUGUAGCUUCUCCGGCUUUAGCUGCCAUUUU ...((((((((((((((.......))))))))).(((....(((.....)))................((((..(..(((.....))).)..)))).....))).....)))))...... ( -29.80) >DroSec_CAF1 18136 120 - 1 UUCGCAGCAGCUUCAUAUUUCAUUUGUGGGGCUACCGUAUUGCGGCGUUCGCUUUUGAUAACUCACCUGCUGUGGCUUAAUCCAUUUAUGUGUAGCUUCUCCGGCUUUAGCUGCCAUUUU ...((((((((((((((.......))))))))).(((.....))).....(((...((...(((((.((..((((......)))).)).))).))..))...)))....)))))...... ( -31.10) >DroSim_CAF1 19276 120 - 1 UUCGCAGCAGCUUCAUAUUUCAUUUGUGGGGCUACCGUAUUGCGGCGUUCGCUUUUGAUAACUCACCUGCUGUGGCUUAAUCCAUUUUUUUGUAGCUUCUCCGGCUUUAGCUGCCAUUUU .((((((((((((((((.......))))))))).(((.....))).......................)))))))................((((((...........))))))...... ( -29.40) >DroEre_CAF1 19157 120 - 1 UUCGCAGCAGCUUCGUAUUUCAUUUGUGGGGCUACCGUAUUUCGGCGUUCGCUUUUGAUAACUCACCUGCUGUGGAUUAAUCCAUUUAUUUGUAGCUUCUCCGGCUUUAGCUGCCAUUUU ...((((((((((((((.......))))))))).(((((((..(((....)))...))))........((((..(((..........)))..)))).....))).....)))))...... ( -28.70) >DroYak_CAF1 18669 120 - 1 UUCGCAGCAGCUUCAUAUUUCAUUUGUGGGGCUACCGUAUUUCGGCGUUCGCUUUUGAUAACUCACCUGCUGUGGCUUAAUCCACUUAUUUGUAGCUUCUCCGGCUUUAGCUGCCAUUUU ...((((((((((((((.......))))))))).(((((((..(((....)))...))))......((((.((((......))))......))))......))).....)))))...... ( -30.90) >consensus UUCGCAGCAGCUUCAUAUUUCAUUUGUGGGGCUACCGUAUUGCGGCGUUCGCUUUUGAUAACUCACCUGCUGUGGCUUAAUCCAUUUAUUUGUAGCUUCUCCGGCUUUAGCUGCCAUUUU ...((((((((((((((.......))))))))).(((((((..(((....)))...))))......((((.((((......))))......))))......))).....)))))...... (-29.24 = -28.92 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:58 2006