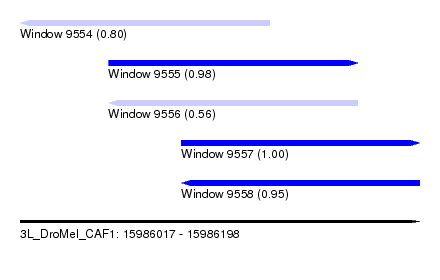
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,986,017 – 15,986,198 |
| Length | 181 |
| Max. P | 0.999654 |

| Location | 15,986,017 – 15,986,130 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.16 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -25.18 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15986017 113 - 23771897 CAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUCGCCG-------GUCGCCGAUAAGCAAUUAAAAUUUCAUUUAAUGCACAACUUUAUUAUCGGUUCGGCCCAGUCACGGUCA .....(((((..((.(((......)))(((((.((((....)))-------)..(((((((((((.(((((......)))))))).........))))))))..)))))))...))))). ( -32.60) >DroSec_CAF1 15058 113 - 1 CAUUCGGCCGCUCUAAGCCCAACAGAUGGGCUCCCGGAUCGCCG-------GUCGCCGAUAAGCAAUUAAAAUUUCAUUUAAUGCACAACUUUAUUAUCGGUUCGGCCCAGUCACGGUCA .....(((((..((.((((((.....))))))...((...((((-------(..(((((((((((.(((((......)))))))).........)))))))))))))))))...))))). ( -36.10) >DroSim_CAF1 16133 113 - 1 CAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUCGCCG-------GUCGCCGAUAAGCAAUUAAAAUUUCAUUUAAUGCACAACUUUAUUAUCGGUUCGGCCCAGUCACGGUCA .....(((((..((.(((......)))(((((.((((....)))-------)..(((((((((((.(((((......)))))))).........))))))))..)))))))...))))). ( -32.60) >DroEre_CAF1 15955 120 - 1 CAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUCACCGAUCGCCGAUCGUCGAUAAGCAAUUAAAAUUUCAUUUAAUGCACAACUUUAUUAUCGGUUUGUCCCAGUCACGGUCA .....(((((..((...((((.....)))).....((((.((((((((.(....).)))...(((.(((((......))))))))............)))))..)))).))...))))). ( -27.40) >DroYak_CAF1 15504 113 - 1 CAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUCGCCG-------GUCGCCGAUAAGCAAUUAAAAUUUCAUUUAAUGCACAACUUUAUUAUCGGUUUGUCCCAGUCACGGUCA .....(((((..((.(((......)))((((..((((....)))-------)..(((((((((((.(((((......)))))))).........))))))))..)))).))...))))). ( -28.20) >consensus CAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUCGCCG_______GUCGCCGAUAAGCAAUUAAAAUUUCAUUUAAUGCACAACUUUAUUAUCGGUUCGGCCCAGUCACGGUCA .....(((((.(((.........)))((((((..(((....)))..........(((((((((((.(((((......)))))))).........))))))))..))))))....))))). (-25.18 = -25.42 + 0.24)


| Location | 15,986,057 – 15,986,170 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.34 |
| Mean single sequence MFE | -44.52 |
| Consensus MFE | -37.56 |
| Energy contribution | -37.16 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15986057 113 + 23771897 AAAUGAAAUUUUAAUUGCUUAUCGGCGAC-------CGGCGAUCCGGGAGCCCAUCUGUUGGGAUUAGAGCGGCCGAAUGGCCGACUCGAUUGAUGUCAAGCGAUCGGUCGGUCGGCGCU ......................((.((((-------(((((((((..((.((((.....))))....((((((((....))))).)))........))..).)))).)))))))).)).. ( -44.10) >DroSec_CAF1 15098 113 + 1 AAAUGAAAUUUUAAUUGCUUAUCGGCGAC-------CGGCGAUCCGGGAGCCCAUCUGUUGGGCUUAGAGCGGCCGAAUGGCCGACUCGAUUGAUGUCAAGCGAUCGGUCGGUCGGCGCU ................((...(((((..(-------(((....))))(((((((.....)))))))......)))))...((((((.((((((((........)))))))))))))))). ( -49.60) >DroSim_CAF1 16173 113 + 1 AAAUGAAAUUUUAAUUGCUUAUCGGCGAC-------CGGCGAUCCGGGAGCCCAUCUGUUGGGAUUAGAGCGGCCGAAUGGCCGACUCGAUUGAUGUCAAGCGAUCGGUCGGUCGGCGCU ......................((.((((-------(((((((((..((.((((.....))))....((((((((....))))).)))........))..).)))).)))))))).)).. ( -44.10) >DroEre_CAF1 15995 120 + 1 AAAUGAAAUUUUAAUUGCUUAUCGACGAUCGGCGAUCGGUGAUCCGGGAGCCCAUCUGUUGGGAUUAGAGCGGCCGAAUGGUCGACUCGAUUGAUGUCAAGCGAUCGGUCGGUCGGCGCU ...............(((((.((((((((.(((..((((....))))..))).))).))))).....)))))(((((.((..(((.(((.(((....))).))))))..)).)))))... ( -44.10) >DroYak_CAF1 15544 113 + 1 AAAUGAAAUUUUAAUUGCUUAUCGGCGAC-------CGGCGAUCCGGGAGCCCAUCUGUUGGGAUUAGAGCGGCCGAAUGGUCGAGUCGAUUGAUGUCAAGCGAUCGGUCGGUCGGCGCU .........(((((((.(.....(((..(-------(((....))))..)))........).)))))))(((.((((.((..(((.(((.(((....))).))))))..)).))))))). ( -40.72) >consensus AAAUGAAAUUUUAAUUGCUUAUCGGCGAC_______CGGCGAUCCGGGAGCCCAUCUGUUGGGAUUAGAGCGGCCGAAUGGCCGACUCGAUUGAUGUCAAGCGAUCGGUCGGUCGGCGCU ...............(((((((((.((.........)).)))).......((((.....))))....)))))(((((.(((((((.(((.(((....))).)))))))))).)))))... (-37.56 = -37.16 + -0.40)


| Location | 15,986,057 – 15,986,170 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.34 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -27.40 |
| Energy contribution | -28.04 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15986057 113 - 23771897 AGCGCCGACCGACCGAUCGCUUGACAUCAAUCGAGUCGGCCAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUCGCCG-------GUCGCCGAUAAGCAAUUAAAAUUUCAUUU ..((.((((((..((((((.(((....))).)(((.(((((....))))))))....((((.....))))......))))).))-------)))).))...................... ( -37.00) >DroSec_CAF1 15098 113 - 1 AGCGCCGACCGACCGAUCGCUUGACAUCAAUCGAGUCGGCCAUUCGGCCGCUCUAAGCCCAACAGAUGGGCUCCCGGAUCGCCG-------GUCGCCGAUAAGCAAUUAAAAUUUCAUUU ..((.((((((..((((((.(((....))).)(((.(((((....))))))))..((((((.....))))))....))))).))-------)))).))...................... ( -41.90) >DroSim_CAF1 16173 113 - 1 AGCGCCGACCGACCGAUCGCUUGACAUCAAUCGAGUCGGCCAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUCGCCG-------GUCGCCGAUAAGCAAUUAAAAUUUCAUUU ..((.((((((..((((((.(((....))).)(((.(((((....))))))))....((((.....))))......))))).))-------)))).))...................... ( -37.00) >DroEre_CAF1 15995 120 - 1 AGCGCCGACCGACCGAUCGCUUGACAUCAAUCGAGUCGACCAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUCACCGAUCGCCGAUCGUCGAUAAGCAAUUAAAAUUUCAUUU .((..((((((.((((((((((((......))))).)))....)))).))..............(((.(((...(((....)))...))).)))))))....))................ ( -31.70) >DroYak_CAF1 15544 113 - 1 AGCGCCGACCGACCGAUCGCUUGACAUCAAUCGACUCGACCAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUCGCCG-------GUCGCCGAUAAGCAAUUAAAAUUUCAUUU ((((((((..(..((((((.(((....))).))).)))..)..)))).))))...........((((((((..((((....)))-------)..)))..(((....)))......))))) ( -31.90) >consensus AGCGCCGACCGACCGAUCGCUUGACAUCAAUCGAGUCGGCCAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUCGCCG_______GUCGCCGAUAAGCAAUUAAAAUUUCAUUU .(((((((((((..(((........)))..))).))))))...(((((.((......((((.....))))....(((....))).......)).)))))...))................ (-27.40 = -28.04 + 0.64)



| Location | 15,986,090 – 15,986,198 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.15 |
| Mean single sequence MFE | -43.66 |
| Consensus MFE | -41.32 |
| Energy contribution | -41.08 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.84 |
| SVM RNA-class probability | 0.999654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15986090 108 + 23771897 GAUCCGGGAGCCCAUCUGUUGGGAUUAGAGCGGCCGAAUGGCCGACUCGAUUGAUGUCAAGCGAUCGGUCGGUCGGCGCUUUGAAGAUUAUGCCCAAAGUUAUGAGGU------------ .....(((..((((.....)))).((((((((.((((.(((((((.(((.(((....))).)))))))))).))))))))))))........))).............------------ ( -43.50) >DroSec_CAF1 15131 108 + 1 GAUCCGGGAGCCCAUCUGUUGGGCUUAGAGCGGCCGAAUGGCCGACUCGAUUGAUGUCAAGCGAUCGGUCGGUCGGCGCUUUGAAGAUUAUGCCCAAAGUUAUGAGGC------------ .....(((.(((((.....)))))((((((((.((((.(((((((.(((.(((....))).)))))))))).))))))))))))........))).............------------ ( -46.80) >DroSim_CAF1 16206 108 + 1 GAUCCGGGAGCCCAUCUGUUGGGAUUAGAGCGGCCGAAUGGCCGACUCGAUUGAUGUCAAGCGAUCGGUCGGUCGGCGCUUUGAAGAUUAUGCCCAAAGUUAUGAGGC------------ .....(((..((((.....)))).((((((((.((((.(((((((.(((.(((....))).)))))))))).))))))))))))........))).............------------ ( -43.50) >DroEre_CAF1 16035 108 + 1 GAUCCGGGAGCCCAUCUGUUGGGAUUAGAGCGGCCGAAUGGUCGACUCGAUUGAUGUCAAGCGAUCGGUCGGUCGGCGCUUUGAAGAUUAUGCCCAAAGUUAUGAGGC------------ .....(((..((((.....)))).((((((((.((((.((..(((.(((.(((....))).))))))..)).))))))))))))........))).............------------ ( -40.30) >DroYak_CAF1 15577 120 + 1 GAUCCGGGAGCCCAUCUGUUGGGAUUAGAGCGGCCGAAUGGUCGAGUCGAUUGAUGUCAAGCGAUCGGUCGGUCGGCGCUUUGAAGAUUAUGCCCAAAGUUAUGAGGCUGAAACUGCGGC ...(((.((((((((...(((((.((((((((.((((.((..(((.(((.(((....))).))))))..)).))))))))))))........)))))....))).)))).....).))). ( -44.20) >consensus GAUCCGGGAGCCCAUCUGUUGGGAUUAGAGCGGCCGAAUGGCCGACUCGAUUGAUGUCAAGCGAUCGGUCGGUCGGCGCUUUGAAGAUUAUGCCCAAAGUUAUGAGGC____________ .....(((..((((.....)))).((((((((.((((.(((((((.(((.(((....))).)))))))))).))))))))))))........)))......................... (-41.32 = -41.08 + -0.24)


| Location | 15,986,090 – 15,986,198 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.15 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -29.42 |
| Energy contribution | -29.82 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15986090 108 - 23771897 ------------ACCUCAUAACUUUGGGCAUAAUCUUCAAAGCGCCGACCGACCGAUCGCUUGACAUCAAUCGAGUCGGCCAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUC ------------..........((((((............((((((((..(.(((((((.(((....))).))).)))).)..)))).)))).....((((.....))))..)))))).. ( -31.40) >DroSec_CAF1 15131 108 - 1 ------------GCCUCAUAACUUUGGGCAUAAUCUUCAAAGCGCCGACCGACCGAUCGCUUGACAUCAAUCGAGUCGGCCAUUCGGCCGCUCUAAGCCCAACAGAUGGGCUCCCGGAUC ------------(((.(((....((((((.......((((.(((.((......))..)))))))........(((.(((((....))))))))...))))))...))))))......... ( -37.80) >DroSim_CAF1 16206 108 - 1 ------------GCCUCAUAACUUUGGGCAUAAUCUUCAAAGCGCCGACCGACCGAUCGCUUGACAUCAAUCGAGUCGGCCAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUC ------------(((.(((....(((((........((((.(((.((......))..)))))))........(((.(((((....))))))))....)))))...))))))......... ( -33.00) >DroEre_CAF1 16035 108 - 1 ------------GCCUCAUAACUUUGGGCAUAAUCUUCAAAGCGCCGACCGACCGAUCGCUUGACAUCAAUCGAGUCGACCAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUC ------------(((.(((....(((((............((((((((..(..((((((.(((....))).))).)))..)..)))).)))).....)))))...))))))......... ( -28.33) >DroYak_CAF1 15577 120 - 1 GCCGCAGUUUCAGCCUCAUAACUUUGGGCAUAAUCUUCAAAGCGCCGACCGACCGAUCGCUUGACAUCAAUCGACUCGACCAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUC .(((.......((((.(((....(((((............((((((((..(..((((((.(((....))).))).)))..)..)))).)))).....)))))...)))))))..)))... ( -30.63) >consensus ____________GCCUCAUAACUUUGGGCAUAAUCUUCAAAGCGCCGACCGACCGAUCGCUUGACAUCAAUCGAGUCGGCCAUUCGGCCGCUCUAAUCCCAACAGAUGGGCUCCCGGAUC ......................((((((............((((((((..(.(((((((.(((....))).))).)))).)..)))).)))).....((((.....))))..)))))).. (-29.42 = -29.82 + 0.40)

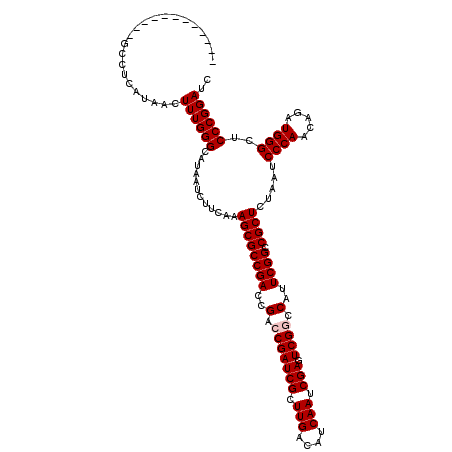
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:49 2006