
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,982,929 – 15,983,289 |
| Length | 360 |
| Max. P | 0.996246 |

| Location | 15,982,929 – 15,983,049 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.95 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -29.10 |
| Energy contribution | -28.86 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15982929 120 + 23771897 UCCUCCGGCUGCAACAAUGGCAUCGUUGUCCCAUAAUCUACACACCACCAUCGGCGGACCUUUUGAAGGUCCUUUGGGCCAAAGUUCUGGCCGUGAUGGCGAACACACACGCCGCCAUAU ......(((.((.((((((....))))))................(.((((((.(((((((.....))))))....(((((......)))))))))))).).........)).))).... ( -39.20) >DroSec_CAF1 12144 106 + 1 UCCUCCGGCUGCACCAAUGGCAUCGCUGUCCCAUAAUCUACACACCACCAUCGGCGGACCUUUUGAAGGUCCUUUGGGCCAAA--------------GGCGAACACACACGCCGCCGCAU .....((((........((((...((((.......................))))((((((.....)))))).....))))..--------------((((........))))))))... ( -32.20) >DroSim_CAF1 12934 106 + 1 UCCUCCGGCUGCAACAAUGGCAUCGUUGUCCCAUAAUCUACACACCACCAUCGGCGGACCUUUUGAAGGUCCUUUGGGCCAAA--------------GGCGAACACACACGCCGCCGCAU .....(((((((.......)))...(((.((((...........((......)).((((((.....))))))..)))).))).--------------((((........))))))))... ( -31.30) >DroEre_CAF1 12802 114 + 1 UCCUCCGGCUGCAACAAUGGCAGCGCUGUCCCAUAAUCUAC--ACCACCAUCGGCGGACCUUUUGAAGGUCCUUUCGGCCAAAGUUCUGGCCGUAAGGGCGAACACACACGCCGCC---- .......(((((.......))))).................--.........(((((((((.....))))))...((((((......))))))....((((........)))))))---- ( -38.30) >DroYak_CAF1 12291 114 + 1 UCCUCCGGCUGCAACAAUGGCAUCGUUGUCCUAUAAUCUAC--ACCACCAGCGGCGGACCUUUUGAAGGUCCUUUGGGCCAAAGUUCUGGCCGUAAGGGCGAACACACACGCCGCC---- ......(((.((.....((((.((((((.............--.....)))))).((((((.....)))))).....))))..((((.(.((....)).)))))......)).)))---- ( -36.67) >consensus UCCUCCGGCUGCAACAAUGGCAUCGUUGUCCCAUAAUCUACACACCACCAUCGGCGGACCUUUUGAAGGUCCUUUGGGCCAAAGUUCUGGCCGU_A_GGCGAACACACACGCCGCC__AU ......(((........((((...((((.......................))))((((((.....)))))).....))))................((((........))))))).... (-29.10 = -28.86 + -0.24)

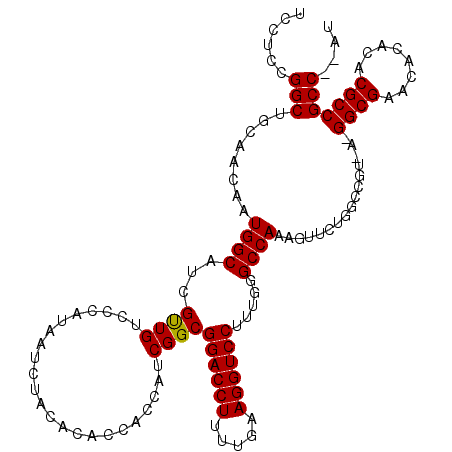
| Location | 15,982,969 – 15,983,089 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.14 |
| Mean single sequence MFE | -45.30 |
| Consensus MFE | -37.66 |
| Energy contribution | -37.70 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15982969 120 - 23771897 UGCGCUUCAAAGAACCUCAUUGUGUUUUGCAUAUGCUGCUAUAUGGCGGCGUGUGUGUUCGCCAUCACGGCCAGAACUUUGGCCCAAAGGACCUUCAAAAGGUCCGCCGAUGGUGGUGUG .((((.....((((((.....).)))))((((((((((((....))))))))))))...(((((((..((((((....))))))....((((((.....))))))...))))))))))). ( -51.90) >DroSec_CAF1 12184 106 - 1 UGCGCUUCAAAGAACCUCAUUGUGUUUUGCAUAUGCUGCUAUGCGGCGGCGUGUGUGUUCGCC--------------UUUGGCCCAAAGGACCUUCAAAAGGUCCGCCGAUGGUGGUGUG .((((.....((((((.....).)))))((((((((((((....))))))))))))...((((--------------.(((((.....((((((.....))))))))))).)))))))). ( -42.60) >DroSim_CAF1 12974 106 - 1 UGCGCUUCAAAGAACCUCAUUGUGUUUUGCAUAUGCUGCUAUGCGGCGGCGUGUGUGUUCGCC--------------UUUGGCCCAAAGGACCUUCAAAAGGUCCGCCGAUGGUGGUGUG .((((.....((((((.....).)))))((((((((((((....))))))))))))...((((--------------.(((((.....((((((.....))))))))))).)))))))). ( -42.60) >DroEre_CAF1 12842 114 - 1 UGCGCUUCAAAGAACCUCAUUGUGUUUUGCAUAUGCUGCU----GGCGGCGUGUGUGUUCGCCCUUACGGCCAGAACUUUGGCCGAAAGGACCUUCAAAAGGUCCGCCGAUGGUGGU--G .............(((((((((......((((((((((..----..))))))))))....((.....(((((((....)))))))...((((((.....)))))))))))))).)))--. ( -44.20) >DroYak_CAF1 12331 114 - 1 UGCGCUUCAAAGAACCUCAUUGUGUUUUGCAUAUGCUGCC----GGCGGCGUGUGUGUUCGCCCUUACGGCCAGAACUUUGGCCCAAAGGACCUUCAAAAGGUCCGCCGCUGGUGGU--G ((((...(((.(.....).))).....))))...((..((----(((((((((......)))......((((((....))))))....((((((.....))))))))))))))..))--. ( -45.20) >consensus UGCGCUUCAAAGAACCUCAUUGUGUUUUGCAUAUGCUGCUAU__GGCGGCGUGUGUGUUCGCC_U_ACGGCCAGAACUUUGGCCCAAAGGACCUUCAAAAGGUCCGCCGAUGGUGGUGUG .............(((((((((((...(((((((((((((....)))))))))))))..)))..................(((.....((((((.....)))))))))))))).)))... (-37.66 = -37.70 + 0.04)

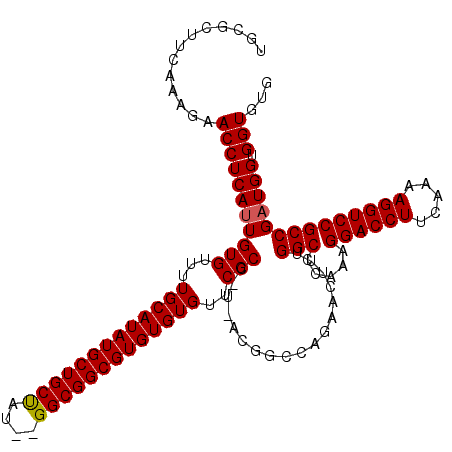

| Location | 15,983,049 – 15,983,169 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.24 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.02 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15983049 120 - 23771897 ACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUAAUUGUCGCUUACCUGAAAGAGCUGAAAGAGGGGAAUGCGAUGCGCUUCAAAGAACCUCAUUGUGUUUUGCAUAUGCUGCU ((....))......((((((((((....((..(((((....))))))))))))))))).(((.(.....(.(((((((((((.(.(((...))).).))))))))))).).....).))) ( -27.50) >DroSec_CAF1 12250 111 - 1 ACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUAAUUGUCGCUUACCUGAA---------GGAGGCGAAUGCGAUGCGCUUCAAAGAACCUCAUUGUGUUUUGCAUAUGCUGCU ((....))......((((((((((....((..(((((....))))))))))))))))---------)..((((.((((((.((((................)))).)))))).))))... ( -28.59) >DroSim_CAF1 13040 111 - 1 ACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUAAUUGUCGCUUACCUGAA---------GGAGGGGAAUGCGAUGCGCUUCAAAGAACCUCAUUGUGUUUUGCAUAUGCUGCU .....(((((....((((((((((....((..(((((....))))))))))))))))---------)..(.(((((((((((.(.(((...))).).))))))))))).)..)))))... ( -25.90) >DroEre_CAF1 12916 110 - 1 ACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUAAUUGUCGC-UACCUGAA---------AGAGGGGAACACGAUGCGCUUCAAAGAACCUCAUUGUGUUUUGCAUAUGCUGCU .....(((((....(((((((((.........(((((....)))))..-))))))))---------)..(.(((((((((((.(.(((...))).).))))))))))).)..)))))... ( -27.90) >DroYak_CAF1 12405 111 - 1 ACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUAAUUGUCGCUUACCUGAA---------AGAGGGGAACACGAUGCGCUUCAAAGAACCUCAUUGUGUUUUGCAUAUGCUGCC .....(((((....((((((((((....((..(((((....))))))))))))))))---------)..(.(((((((((((.(.(((...))).).))))))))))).)..)))))... ( -29.10) >consensus ACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUAAUUGUCGCUUACCUGAA_________AGAGGGGAAUGCGAUGCGCUUCAAAGAACCUCAUUGUGUUUUGCAUAUGCUGCU .....(((((.....(((((((((....((..(((((....))))))))))))))))............(.(((((((((((.(.(((...))).).))))))))))).)..)))))... (-25.10 = -25.02 + -0.08)



| Location | 15,983,089 – 15,983,209 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -19.27 |
| Energy contribution | -18.99 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15983089 120 + 23771897 UCGCAUUCCCCUCUUUCAGCUCUUUCAGGUAAGCGACAAUUAUUUAUCUUACAAAUUUACCUGAAAUUAAAUACUUUCGUUUGACGGCGCUGCUGCGCAAUGCAGCUCUACACAAAAGGC .........(((......(((.(((((((((((......................)))))))))))((((((......)))))).)))(((((........)))))..........))). ( -23.35) >DroSec_CAF1 12290 111 + 1 UCGCAUUCGCCUCC---------UUCAGGUAAGCGACAAUUAUUUAUCUUACAAAUUUACCUGAAAUUAAAUACUUUCGUUUGACGGCGCUGCUGCGCAAUGCAGCGCUACACAAAAGGC ........((((..---------((((((((((......................))))))))))..............((((..((((((((........))))))))...)))))))) ( -30.05) >DroSim_CAF1 13080 111 + 1 UCGCAUUCCCCUCC---------UUCAGGUAAGCGACAAUUAUUUAUCUUACAAAUUUACCUGAAAUUAAAUACUUUCGUUUGACGGCGCUGCUGCGCAAUGCAGCGCUACACAAAAGGC .........(((..---------((((((((((......................))))))))))..............((((..((((((((........))))))))...))))))). ( -27.05) >DroEre_CAF1 12956 105 + 1 UCGUGUUCCCCUCU---------UUCAGGUA-GCGACAAUUAUUUAUCUUACAAAUUUACCUGAAAUUAAAUACUUUCGUUUGACGGCGC---UGCGCAAUGCAG--CUACACAAAAGGC ..((((...((..(---------((((((((-(.......................))))))))))((((((......)))))).)).((---(((.....))))--).))))....... ( -22.10) >DroYak_CAF1 12445 106 + 1 UCGUGUUCCCCUCU---------UUCAGGUAAGCGACAAUUAUUUAUCUUACAAAUUUACCUGAAAUUAAAUACUUUCGUUUGACGGCGC---UGCGCAAUGCAG--CUACACAAAAGGC ..((((...((..(---------((((((((((......................)))))))))))((((((......)))))).)).((---(((.....))))--).))))....... ( -22.95) >consensus UCGCAUUCCCCUCU_________UUCAGGUAAGCGACAAUUAUUUAUCUUACAAAUUUACCUGAAAUUAAAUACUUUCGUUUGACGGCGCUGCUGCGCAAUGCAGC_CUACACAAAAGGC ..(((((................((((((((((......................)))))))))).((((((......))))))..((((....)))))))))................. (-19.27 = -18.99 + -0.28)


| Location | 15,983,089 – 15,983,209 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -25.36 |
| Energy contribution | -25.78 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.937990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15983089 120 - 23771897 GCCUUUUGUGUAGAGCUGCAUUGCGCAGCAGCGCCGUCAAACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUAAUUGUCGCUUACCUGAAAGAGCUGAAAGAGGGGAAUGCGA .((((((.......(((((.....)))))(((........((....))......((((((((((....((..(((((....)))))))))))))))))..)))..))))))......... ( -32.20) >DroSec_CAF1 12290 111 - 1 GCCUUUUGUGUAGCGCUGCAUUGCGCAGCAGCGCCGUCAAACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUAAUUGUCGCUUACCUGAA---------GGAGGCGAAUGCGA ((((((((.(..(((((((........)))))))..))))((....))......((((((((((....((..(((((....))))))))))))))))---------))))))........ ( -35.40) >DroSim_CAF1 13080 111 - 1 GCCUUUUGUGUAGCGCUGCAUUGCGCAGCAGCGCCGUCAAACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUAAUUGUCGCUUACCUGAA---------GGAGGGGAAUGCGA ...((((((...(((((((........)))))))......))))))((((((..((((((((((....((..(((((....))))))))))))))))---------).....)))))).. ( -33.40) >DroEre_CAF1 12956 105 - 1 GCCUUUUGUGUAG--CUGCAUUGCGCA---GCGCCGUCAAACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUAAUUGUCGC-UACCUGAA---------AGAGGGGAACACGA .....((((((.(--((((.....)))---)).((.((..((....))......(((((((((.........(((((....)))))..-))))))))---------))).))..)))))) ( -32.40) >DroYak_CAF1 12445 106 - 1 GCCUUUUGUGUAG--CUGCAUUGCGCA---GCGCCGUCAAACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUAAUUGUCGCUUACCUGAA---------AGAGGGGAACACGA .....((((((.(--((((.....)))---)).((.((..((....))......((((((((((....((..(((((....))))))))))))))))---------))).))..)))))) ( -33.60) >consensus GCCUUUUGUGUAG_GCUGCAUUGCGCAGCAGCGCCGUCAAACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUAAUUGUCGCUUACCUGAA_________AGAGGGGAAUGCGA .....((((((...(((((.....)))))....((.((..((....)).......(((((((((....((..(((((....))))))))))))))))..........)).))..)))))) (-25.36 = -25.78 + 0.42)



| Location | 15,983,129 – 15,983,249 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -24.80 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15983129 120 + 23771897 UAUUUAUCUUACAAAUUUACCUGAAAUUAAAUACUUUCGUUUGACGGCGCUGCUGCGCAAUGCAGCUCUACACAAAAGGCGACAAAUUCCCCCUUUUUUGCCAUGUGUGCGUCGUGUUUU ...........(((((......((((........)))))))))(((((((.(((((.....)))))...(((((...(((((.(((.......))).))))).))))))))))))..... ( -29.70) >DroSec_CAF1 12321 119 + 1 UAUUUAUCUUACAAAUUUACCUGAAAUUAAAUACUUUCGUUUGACGGCGCUGCUGCGCAAUGCAGCGCUACACAAAAGGCGACAAAUUCCCUCUU-UUUGCCAUGUGUGCGUCGUGUUUU .........(((..........((((........))))....(((((((((((........)))))))((((((...(((((.((........))-.))))).))))))))))))).... ( -31.30) >DroSim_CAF1 13111 119 + 1 UAUUUAUCUUACAAAUUUACCUGAAAUUAAAUACUUUCGUUUGACGGCGCUGCUGCGCAAUGCAGCGCUACACAAAAGGCGACAAAUUCCCCCUU-UUUGCCAUGUGUGCGUCGUGUUUU .........(((..........((((........))))....(((((((((((........)))))))((((((...(((((.((........))-.))))).))))))))))))).... ( -31.30) >DroEre_CAF1 12986 114 + 1 UAUUUAUCUUACAAAUUUACCUGAAAUUAAAUACUUUCGUUUGACGGCGC---UGCGCAAUGCAG--CUACACAAAAGGCGACAAAUUCCCCCUU-UUUGCCAUGUGUGCGUCGUGUUUU ...........(((((......((((........)))))))))(((((((---(((.....))))--)((((((...(((((.((........))-.))))).)))))).)))))..... ( -25.30) >DroYak_CAF1 12476 114 + 1 UAUUUAUCUUACAAAUUUACCUGAAAUUAAAUACUUUCGUUUGACGGCGC---UGCGCAAUGCAG--CUACACAAAAGGCGACAAAUUCCCCCUU-UUUGCCAUGUGUGCGUCGUGUUUU ...........(((((......((((........)))))))))(((((((---(((.....))))--)((((((...(((((.((........))-.))))).)))))).)))))..... ( -25.30) >consensus UAUUUAUCUUACAAAUUUACCUGAAAUUAAAUACUUUCGUUUGACGGCGCUGCUGCGCAAUGCAGC_CUACACAAAAGGCGACAAAUUCCCCCUU_UUUGCCAUGUGUGCGUCGUGUUUU ...........(((((......((((........)))))))))(((((((.(((((.....)))))...(((((...(((((...............))))).))))))))))))..... (-24.80 = -25.30 + 0.50)


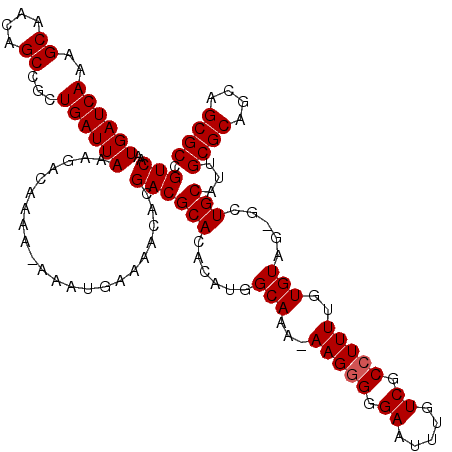
| Location | 15,983,129 – 15,983,249 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -24.24 |
| Energy contribution | -24.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15983129 120 - 23771897 AAAACACGACGCACACAUGGCAAAAAAGGGGGAAUUUGUCGCCUUUUGUGUAGAGCUGCAUUGCGCAGCAGCGCCGUCAAACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUA .....(((.(((.......(((.((((((.((......)).)))))).)))...(((((.....))))).))).)))...(((((..(((((......)))))..))))).......... ( -29.20) >DroSec_CAF1 12321 119 - 1 AAAACACGACGCACACAUGGCAAA-AAGAGGGAAUUUGUCGCCUUUUGUGUAGCGCUGCAUUGCGCAGCAGCGCCGUCAAACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUA .......((((.(((((.(((..(-((.......)))...)))...))))).(((((((........)))))))))))..(((((..(((((......)))))..))))).......... ( -31.20) >DroSim_CAF1 13111 119 - 1 AAAACACGACGCACACAUGGCAAA-AAGGGGGAAUUUGUCGCCUUUUGUGUAGCGCUGCAUUGCGCAGCAGCGCCGUCAAACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUA .......((((........(((.(-(((((.((.....)).)))))).))).(((((((........)))))))))))..(((((..(((((......)))))..))))).......... ( -32.40) >DroEre_CAF1 12986 114 - 1 AAAACACGACGCACACAUGGCAAA-AAGGGGGAAUUUGUCGCCUUUUGUGUAG--CUGCAUUGCGCA---GCGCCGUCAAACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUA .......(((((.......(((.(-(((((.((.....)).)))))).))).(--((((.....)))---))).))))..(((((..(((((......)))))..))))).......... ( -28.40) >DroYak_CAF1 12476 114 - 1 AAAACACGACGCACACAUGGCAAA-AAGGGGGAAUUUGUCGCCUUUUGUGUAG--CUGCAUUGCGCA---GCGCCGUCAAACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUA .......(((((.......(((.(-(((((.((.....)).)))))).))).(--((((.....)))---))).))))..(((((..(((((......)))))..))))).......... ( -28.40) >consensus AAAACACGACGCACACAUGGCAAA_AAGGGGGAAUUUGUCGCCUUUUGUGUAG_GCUGCAUUGCGCAGCAGCGCCGUCAAACGAAAGUAUUUAAUUUCAGGUAAAUUUGUAAGAUAAAUA .......((((((......(((...(((((.((.....)).)))))..))).....)))...((((....)))).)))..(((((..(((((......)))))..))))).......... (-24.24 = -24.44 + 0.20)



| Location | 15,983,169 – 15,983,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.77 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -28.60 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15983169 120 + 23771897 UUGACGGCGCUGCUGCGCAAUGCAGCUCUACACAAAAGGCGACAAAUUCCCCCUUUUUUGCCAUGUGUGCGUCGUGUUUUCAUUUUUUUUGUCUUUAAUCAGCGGCUGUUGCUUUGAUCA ...(((((((.(((((.....)))))...(((((...(((((.(((.......))).))))).))))))))))))......................(((((.(((....)))))))).. ( -33.50) >DroSec_CAF1 12361 118 + 1 UUGACGGCGCUGCUGCGCAAUGCAGCGCUACACAAAAGGCGACAAAUUCCCUCUU-UUUGCCAUGUGUGCGUCGUGUUUUCAUUU-UUUUGUCUUUAAUCAGCGGCUGUUGCUUUGAUCA ..(((((((((((........)))))))((((((...(((((.((........))-.))))).))))))))))............-...........(((((.(((....)))))))).. ( -34.50) >DroSim_CAF1 13151 118 + 1 UUGACGGCGCUGCUGCGCAAUGCAGCGCUACACAAAAGGCGACAAAUUCCCCCUU-UUUGCCAUGUGUGCGUCGUGUUUUCAUUU-UUUUGUCUUUAAUCAGCGGCUGUUGCUUUGAUCA ..(((((((((((........)))))))((((((...(((((.((........))-.))))).))))))))))............-...........(((((.(((....)))))))).. ( -34.50) >DroEre_CAF1 13026 112 + 1 UUGACGGCGC---UGCGCAAUGCAG--CUACACAAAAGGCGACAAAUUCCCCCUU-UUUGCCAUGUGUGCGUCGUGUUUUCAUUU--UUUGUCUUUAAUCAGCGGCUGUUGCUUUGAUCA ...(((((((---(((.....))))--)((((((...(((((.((........))-.))))).)))))).)))))..........--..........(((((.(((....)))))))).. ( -29.10) >DroYak_CAF1 12516 112 + 1 UUGACGGCGC---UGCGCAAUGCAG--CUACACAAAAGGCGACAAAUUCCCCCUU-UUUGCCAUGUGUGCGUCGUGUUUUCAUUU--UUUGUCUUUAAUCAGCGGCUGUUGCUUUGAUCA ...(((((((---(((.....))))--)((((((...(((((.((........))-.))))).)))))).)))))..........--..........(((((.(((....)))))))).. ( -29.10) >consensus UUGACGGCGCUGCUGCGCAAUGCAGC_CUACACAAAAGGCGACAAAUUCCCCCUU_UUUGCCAUGUGUGCGUCGUGUUUUCAUUU_UUUUGUCUUUAAUCAGCGGCUGUUGCUUUGAUCA ...(((((((.(((((.....)))))...(((((...(((((...............))))).))))))))))))......................(((((.(((....)))))))).. (-28.60 = -29.10 + 0.50)



| Location | 15,983,169 – 15,983,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.77 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15983169 120 - 23771897 UGAUCAAAGCAACAGCCGCUGAUUAAAGACAAAAAAAAUGAAAACACGACGCACACAUGGCAAAAAAGGGGGAAUUUGUCGCCUUUUGUGUAGAGCUGCAUUGCGCAGCAGCGCCGUCAA ((((((..((....))...))))))....................(((.(((.......(((.((((((.((......)).)))))).)))...(((((.....))))).))).)))... ( -30.20) >DroSec_CAF1 12361 118 - 1 UGAUCAAAGCAACAGCCGCUGAUUAAAGACAAAA-AAAUGAAAACACGACGCACACAUGGCAAA-AAGAGGGAAUUUGUCGCCUUUUGUGUAGCGCUGCAUUGCGCAGCAGCGCCGUCAA ((((((..((....))...))))))..(((....-........((((...((.(....)))...-((((((((.....)).)))))))))).(((((((........))))))).))).. ( -32.60) >DroSim_CAF1 13151 118 - 1 UGAUCAAAGCAACAGCCGCUGAUUAAAGACAAAA-AAAUGAAAACACGACGCACACAUGGCAAA-AAGGGGGAAUUUGUCGCCUUUUGUGUAGCGCUGCAUUGCGCAGCAGCGCCGUCAA ((((((..((....))...))))))..(((....-........((((...((.(....)))..(-(((((.((.....)).)))))))))).(((((((........))))))).))).. ( -34.40) >DroEre_CAF1 13026 112 - 1 UGAUCAAAGCAACAGCCGCUGAUUAAAGACAAA--AAAUGAAAACACGACGCACACAUGGCAAA-AAGGGGGAAUUUGUCGCCUUUUGUGUAG--CUGCAUUGCGCA---GCGCCGUCAA ((((((..((....))...))))))..(((...--........((((...((.(....)))..(-(((((.((.....)).)))))))))).(--((((.....)))---))...))).. ( -29.50) >DroYak_CAF1 12516 112 - 1 UGAUCAAAGCAACAGCCGCUGAUUAAAGACAAA--AAAUGAAAACACGACGCACACAUGGCAAA-AAGGGGGAAUUUGUCGCCUUUUGUGUAG--CUGCAUUGCGCA---GCGCCGUCAA ((((((..((....))...))))))..(((...--........((((...((.(....)))..(-(((((.((.....)).)))))))))).(--((((.....)))---))...))).. ( -29.50) >consensus UGAUCAAAGCAACAGCCGCUGAUUAAAGACAAAA_AAAUGAAAACACGACGCACACAUGGCAAA_AAGGGGGAAUUUGUCGCCUUUUGUGUAG_GCUGCAUUGCGCAGCAGCGCCGUCAA ((((((..((....))...))))))......................((((((......(((...(((((.((.....)).)))))..))).....)))...((((....)))).))).. (-25.24 = -25.44 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:38 2006