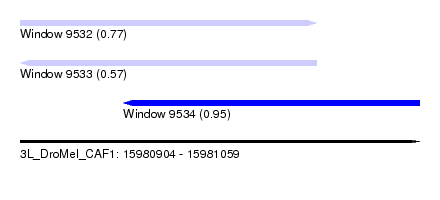
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,980,904 – 15,981,059 |
| Length | 155 |
| Max. P | 0.954070 |

| Location | 15,980,904 – 15,981,019 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -36.96 |
| Consensus MFE | -29.08 |
| Energy contribution | -29.56 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15980904 115 + 23771897 GUUGCGCUUAGUAGCUUAGAAUCAAAGUCGGAUCAGCGCAAAAUUCUGUGAUCUUCUCUGAUCUCCAGCGAUUGCUCUCUGCCGACGUCGCUGCCGCU-----GCCGCUGCUCUGCAGCU ((((((...((((((...........(((((...((.((((....(((.((((......))))..)))...)))).))...)))))((.((....)).-----)).)))))).)))))). ( -37.50) >DroSec_CAF1 10060 109 + 1 GUUGCGCUUAGUAGCUUAGAAUCAAAGUCGGAUCAGCGCAAAAUUCUGUGAUCUUCUCUGAUCUCCAGCGAUUGCGCUCUGCCGACGUCGCUGC-----------CGCUGCUCUGCCGCU ...((((..((((((.(((.......(((((...(((((((....(((.((((......))))..)))...)))))))...)))))....))).-----------.))))))..).))). ( -39.60) >DroSim_CAF1 10773 115 + 1 GUUGCGCUUAGUAGCUUAGAAUCAAAGUCGGAUCAGCGCAAAAUUCUGUGAUCUUCUCUGAUCUCCAGCGAUUGCGCUCUGCCGACGUCGCUGCCGCU-----GCCGCUGCUCUGCCGCU ...((((..((((((...........(((((...(((((((....(((.((((......))))..)))...)))))))...)))))((.((....)).-----)).))))))..).))). ( -41.90) >DroEre_CAF1 10788 120 + 1 GUUGCGCUUAGUAGCUUAGAAUCAAAGUCGGAUCAGCGCAAAUUUCUGUGAUCUUCUCUGAUCUCCAGCGAUUGCUCUCUGCCGACGUCGCUGCCGCUGUUCUGCCGCUGCUCUGCCGCU ...((((..((((((.((((((....(((((...((.((((.((.(((.((((......))))..))).)))))).))...)))))...((....)).))))))..))))))..).))). ( -39.10) >DroYak_CAF1 10313 97 + 1 GUUGCGUUUAGUAGCUUAGAAUCAAAGUCGGAUCAGCGCAAAUUUCUGUGAUCUUCUCAGAUCUCCAGCGAUUGCUCUCUGCCGGC-----------------------GCUCUGCCGCU (((((.....)))))..........((.(((...(((((.((((.(((.(((((....)))))..))).))))((.....))..))-----------------------)))...))))) ( -26.70) >consensus GUUGCGCUUAGUAGCUUAGAAUCAAAGUCGGAUCAGCGCAAAAUUCUGUGAUCUUCUCUGAUCUCCAGCGAUUGCUCUCUGCCGACGUCGCUGCCGCU_____GCCGCUGCUCUGCCGCU ...((((..((((((...........(((((...((.((((....(((.((((......))))..)))...)))).))...))))).........((......)).))))))..)).)). (-29.08 = -29.56 + 0.48)

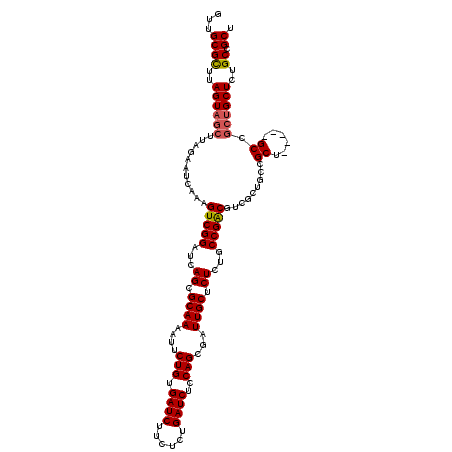
| Location | 15,980,904 – 15,981,019 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -29.60 |
| Energy contribution | -30.30 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15980904 115 - 23771897 AGCUGCAGAGCAGCGGC-----AGCGGCAGCGACGUCGGCAGAGAGCAAUCGCUGGAGAUCAGAGAAGAUCACAGAAUUUUGCGCUGAUCCGACUUUGAUUCUAAGCUACUAAGCGCAAC .(((((......)))))-----.((((...(((.((((((((...((((...(((..((((......)))).)))....)))).)))..))))).)))...))..(((....)))))... ( -38.00) >DroSec_CAF1 10060 109 - 1 AGCGGCAGAGCAGCG-----------GCAGCGACGUCGGCAGAGCGCAAUCGCUGGAGAUCAGAGAAGAUCACAGAAUUUUGCGCUGAUCCGACUUUGAUUCUAAGCUACUAAGCGCAAC .(((.(..((.(((.-----------....(((.(((((...(((((((...(((..((((......)))).)))....)))))))...))))).))).......))).))..))))... ( -38.60) >DroSim_CAF1 10773 115 - 1 AGCGGCAGAGCAGCGGC-----AGCGGCAGCGACGUCGGCAGAGCGCAAUCGCUGGAGAUCAGAGAAGAUCACAGAAUUUUGCGCUGAUCCGACUUUGAUUCUAAGCUACUAAGCGCAAC .((.((......)).))-----.((((...(((.(((((...(((((((...(((..((((......)))).)))....)))))))...))))).)))...))..(((....)))))... ( -40.20) >DroEre_CAF1 10788 120 - 1 AGCGGCAGAGCAGCGGCAGAACAGCGGCAGCGACGUCGGCAGAGAGCAAUCGCUGGAGAUCAGAGAAGAUCACAGAAAUUUGCGCUGAUCCGACUUUGAUUCUAAGCUACUAAGCGCAAC .(((..(((((.((.((......)).))..(((.((((((((...((((...(((..((((......)))).)))....)))).)))..))))).))).......))).))...)))... ( -38.70) >DroYak_CAF1 10313 97 - 1 AGCGGCAGAGC-----------------------GCCGGCAGAGAGCAAUCGCUGGAGAUCUGAGAAGAUCACAGAAAUUUGCGCUGAUCCGACUUUGAUUCUAAGCUACUAAACGCAAC .(((..(((((-----------------------..((((((...((((...(((..(((((....))))).)))....)))).)))..)))...(((....)))))).))...)))... ( -26.60) >consensus AGCGGCAGAGCAGCGGC_____AGCGGCAGCGACGUCGGCAGAGAGCAAUCGCUGGAGAUCAGAGAAGAUCACAGAAUUUUGCGCUGAUCCGACUUUGAUUCUAAGCUACUAAGCGCAAC .(((..(((((.((.((......)).))......((((((((...((((...(((..((((......)))).)))....)))).)))..)))))...........))).))...)))... (-29.60 = -30.30 + 0.70)

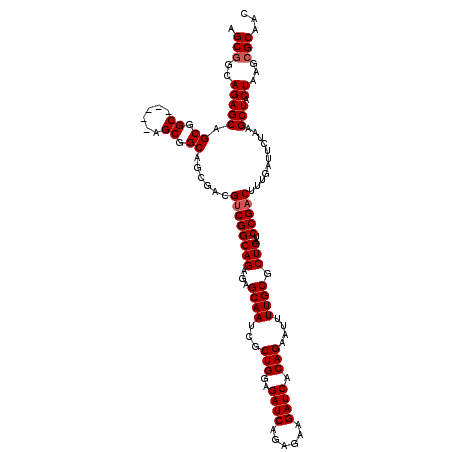
| Location | 15,980,944 – 15,981,059 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -33.16 |
| Consensus MFE | -26.62 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
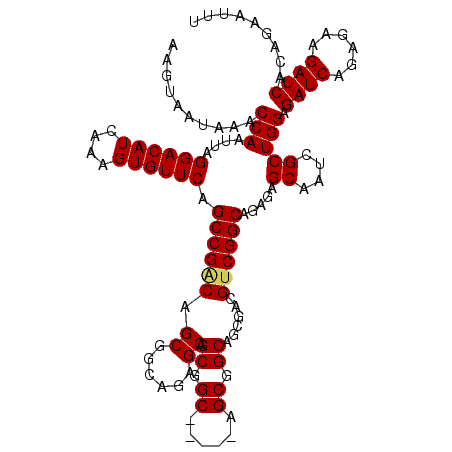
>3L_DroMel_CAF1 15980944 115 - 23771897 AAGUAAUAAACCAAUUAGGACAUCAAAGUGUUCAGCCGGCAGCUGCAGAGCAGCGGC-----AGCGGCAGCGACGUCGGCAGAGAGCAAUCGCUGGAGAUCAGAGAAGAUCACAGAAUUU .................((((((....)))))).((((((.(((((...((......-----.)).)))))...))))))...((....)).(((..((((......)))).)))..... ( -35.90) >DroSec_CAF1 10100 109 - 1 AAGUAAUAAACCAAUUAGGACAUCAAAGUGUUCAGCCGACAGCGGCAGAGCAGCG-----------GCAGCGACGUCGGCAGAGCGCAAUCGCUGGAGAUCAGAGAAGAUCACAGAAUUU ..........((.....))........((((((.((((((.((......)).((.-----------...))...)))))).)))))).....(((..((((......)))).)))..... ( -32.80) >DroSim_CAF1 10813 115 - 1 AAGUAAUAAACCAAUUAGGACAUCAAAGUGUUCAGCCGACAGCGGCAGAGCAGCGGC-----AGCGGCAGCGACGUCGGCAGAGCGCAAUCGCUGGAGAUCAGAGAAGAUCACAGAAUUU ..........((.....))........((((((.((((((.((.((......)).))-----.((....))...)))))).)))))).....(((..((((......)))).)))..... ( -36.90) >DroEre_CAF1 10828 120 - 1 AAGUAAUAAACCAAUUAGGACAUCAAAGUGUUCAGCCGACAGCGGCAGAGCAGCGGCAGAACAGCGGCAGCGACGUCGGCAGAGAGCAAUCGCUGGAGAUCAGAGAAGAUCACAGAAAUU .................((((((....)))))).((((((.((......)).((.((......)).))......))))))...((....)).(((..((((......)))).)))..... ( -34.30) >DroYak_CAF1 10353 97 - 1 AAGUAAUAAACCAAUUAGGACAUCAAAGUGUUCAGCCGACAGCGGCAGAGC-----------------------GCCGGCAGAGAGCAAUCGCUGGAGAUCUGAGAAGAUCACAGAAAUU ..((.............((((((....)))))).((((....))))...))-----------------------.(((((.((......))))))).(((((....)))))......... ( -25.90) >consensus AAGUAAUAAACCAAUUAGGACAUCAAAGUGUUCAGCCGACAGCGGCAGAGCAGCGGC_____AGCGGCAGCGACGUCGGCAGAGAGCAAUCGCUGGAGAUCAGAGAAGAUCACAGAAUUU ..........(((....((((((....)))))).((((((.((......)).((.((......)).))......)))))).....((....))))).((((......))))......... (-26.62 = -27.00 + 0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:26 2006