
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,979,523 – 15,979,653 |
| Length | 130 |
| Max. P | 0.623453 |

| Location | 15,979,523 – 15,979,631 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.86 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -26.91 |
| Energy contribution | -27.91 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15979523 108 - 23771897 CCACGGCACGUGGUAAACGCGCUGCAUCAAAGGCGCCGGCCACAUCCCACCUAAAUGUAAAGCUGGCUUCAAUCUGGCGGCAUC------------CAACAUCCAGCAAGAUUGAAGAUC ...((((..(((((....(((((........)))))..))))).....((......))...)))).(((((((((.((((.((.------------....)))).)).)))))))))... ( -33.80) >DroSec_CAF1 8736 108 - 1 CCACGGCACGUGGUAAACGCGCUGCAUCAAAGGCGCCGGCCACAUCCCACCUCAAUGUAAAGCUGGCUUCAAUCUGGCGGCAUC------------CAACAUCCAGCAAGAUUGAAGAUC ...((((..(((((....(((((........)))))..))))).....((......))...)))).(((((((((.((((.((.------------....)))).)).)))))))))... ( -33.80) >DroSim_CAF1 9411 108 - 1 CCACGGCACGUGGUAGACGCGCUGCAUCAAAGGCGCCGGCCACAUCCCACCUAAAUGUAAAGCUGGCUUCAAUCUGGCGGCAUC------------CAACAUGCAGCAAGAUUGAAGAUC ...((((..(((((....(((((........)))))..))))).....((......))...)))).(((((((((.((.((((.------------....)))).)).)))))))))... ( -38.40) >DroEre_CAF1 8936 120 - 1 CCACGGCACGUGGUAAACGCGCCGCUUCAAAGGCGCCGGCCACAUCCCACCUAAAUGUAAAGCUGGCUUCAGACUGGCGGCAUCGGAAACUCCAGCCAACGUACACAAAGAUCGAAGAUC ....(((..(((.....)))((((((.....((.((((((.((((.........))))...)))))).)).....))))))...((.....)).)))............((((...)))) ( -34.40) >DroYak_CAF1 8849 117 - 1 CCACGGCACGUGGUAAACGCGCUGCUUCAAAGGCGCCGGCCACAUCCCACCUAAAUGUAAGGCUGGCUUCAGUCUGGCGGCAUCGGAGACUCCGACCAACCUCCACCAAGAUCCAAG--- ....((....((((......((((((....(((((((((((((((.........))))..)))))))....)))))))))).((((.....)))).........))))....))...--- ( -36.90) >consensus CCACGGCACGUGGUAAACGCGCUGCAUCAAAGGCGCCGGCCACAUCCCACCUAAAUGUAAAGCUGGCUUCAAUCUGGCGGCAUC____________CAACAUCCAGCAAGAUUGAAGAUC ...((((..(((((....(((((........)))))..))))).....((......))...)))).(((((((((.((...........................)).)))))))))... (-26.91 = -27.91 + 1.00)

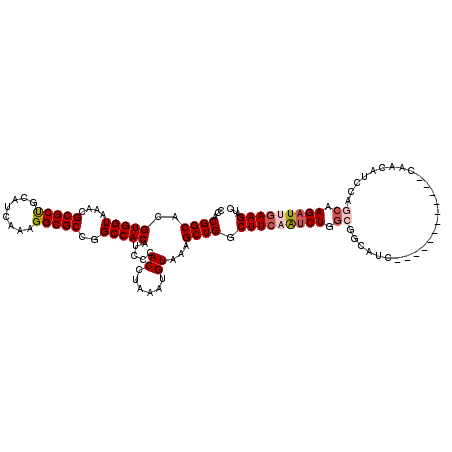

| Location | 15,979,551 – 15,979,653 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.83 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -26.76 |
| Energy contribution | -26.60 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15979551 102 - 23771897 AUGG---------------C---GGAGUGGCUCCAACUUGCCACGGCACGUGGUAAACGCGCUGCAUCAAAGGCGCCGGCCACAUCCCACCUAAAUGUAAAGCUGGCUUCAAUCUGGCGG ..((---------------.---(((((((((.....((((((((...))))))))..(((((........))))).)))))).)))..))..........((..(.......)..)).. ( -35.50) >DroSec_CAF1 8764 83 - 1 AU-------------------------------------GCCACGGCACGUGGUAAACGCGCUGCAUCAAAGGCGCCGGCCACAUCCCACCUCAAUGUAAAGCUGGCUUCAAUCUGGCGG .(-------------------------------------((((((...)))))))..((((((........)))((((((.((((.........))))...)))))).........))). ( -26.70) >DroSim_CAF1 9439 83 - 1 AU-------------------------------------GCCACGGCACGUGGUAGACGCGCUGCAUCAAAGGCGCCGGCCACAUCCCACCUAAAUGUAAAGCUGGCUUCAAUCUGGCGG .(-------------------------------------((((((...)))))))..((((((........)))((((((.((((.........))))...)))))).........))). ( -26.70) >DroEre_CAF1 8976 117 - 1 AUGGUG---GAGGCGGAGGCGGAGGAGUGGCUUGAACUUGCCACGGCACGUGGUAAACGCGCCGCUUCAAAGGCGCCGGCCACAUCCCACCUAAAUGUAAAGCUGGCUUCAGACUGGCGG ..((((---(..(.((.((((..((((((((.((...((((((((...)))))))).)).)))))))).....))))..)).)...)))))..........((..((....).)..)).. ( -45.90) >DroYak_CAF1 8886 117 - 1 AAGGGAUGGGAUGGGAAGGC---GGAGUGGUUUGAACUUGCCACGGCACGUGGUAAACGCGCUGCUUCAAAGGCGCCGGCCACAUCCCACCUAAAUGUAAGGCUGGCUUCAGUCUGGCGG ..(((.(((((((((..(((---(..(((((........)))))((((((((.....)))).)))).......))))..)).))))))))))...(((.((((((....)))))).))). ( -52.80) >consensus AUGG_______________C___GGAGUGG_U__AACUUGCCACGGCACGUGGUAAACGCGCUGCAUCAAAGGCGCCGGCCACAUCCCACCUAAAUGUAAAGCUGGCUUCAAUCUGGCGG .......................................((((((((.(((.....))).)))).......((.((((((.((((.........))))...)))))).))....)))).. (-26.76 = -26.60 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:22 2006