
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,979,322 – 15,979,478 |
| Length | 156 |
| Max. P | 0.996177 |

| Location | 15,979,322 – 15,979,442 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -27.02 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15979322 120 + 23771897 CAAAAUAAAAUACCAUUCAAAGUGGCUGCGAGUUGUUUUUAUCUCGUUAGAGUUGCCAGCGCGCCGAUUUCUUUUCGACUUAAAUGGUUUAUGUGCGUUUGCAACCAGUUUUAUGGACAU ............((((..(((.((.(((((((.((....)).)))).))).(((((.((((((((((.......)))(((.....)))....))))))).))))))).))).)))).... ( -30.10) >DroSec_CAF1 8524 120 + 1 CAAAAUAAAAUACCAUUCAAAGUGGCUGCGAGUUGUUUUUAUCUCGUUAGAGUUGCCAGCGCGCCGAUUUCUUUUCGGCUUAAAUGGUUUAUGUGCGUUUGUAACCAGUUUUAUGGACAU ............((((..(((.((.(((((((.((....)).)))).))).(((((.((((((((((.......))))).(((.....)))...))))).))))))).))).)))).... ( -29.60) >DroSim_CAF1 9206 120 + 1 CAAAAUAAAAUACCAUUCAAAGUGGCUGCAAGUUGUUUUUAUCUCGUUAGAGUUGCCAGCGCGCCGAUUUCUUUUCGGCUUAAAUGGUUUAUGUGCGUUUGCAACCAGUUUUAUGGACAU ............((((..(((.(((.((((((..........(((....)))..((((....(((((.......))))).....)))).........)))))).))).))).)))).... ( -29.90) >DroEre_CAF1 8711 118 + 1 CAAAAUAAAAUACCAUUCAAAGUGGCUCCGAGUUGUU--UAUCUCGUUAGAGUUGCCAGCGCGCCGAUUUCUUUUCGGCUUAAAUGGUUUAUGUGCGUUUGCAACCAGUUUUAUGGACAU ............((((..(((.((((((((((.....--...))))...)))((((.((((((((((.......))))).(((.....)))...))))).))))))).))).)))).... ( -30.60) >DroYak_CAF1 8649 118 + 1 CAAAAUAAAAUACCAUUCAAAGUGGCUCCGAGUUGUU--UAUCUCGUUAGAGUUGCCAGCGCGCCGAUUUCUUUUCGGCUUAAAUGGUUUAUGUGCGUUUGCAACCAGUUUUAUGGACAU ............((((..(((.((((((((((.....--...))))...)))((((.((((((((((.......))))).(((.....)))...))))).))))))).))).)))).... ( -30.60) >consensus CAAAAUAAAAUACCAUUCAAAGUGGCUGCGAGUUGUUUUUAUCUCGUUAGAGUUGCCAGCGCGCCGAUUUCUUUUCGGCUUAAAUGGUUUAUGUGCGUUUGCAACCAGUUUUAUGGACAU ............((((..(((.((.(((((((.((....)).)))).))).(((((.((((((((((.......)))(((.....)))....))))))).))))))).))).)))).... (-27.02 = -27.30 + 0.28)

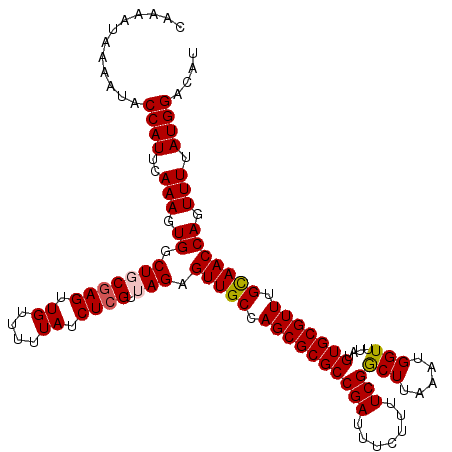

| Location | 15,979,322 – 15,979,442 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -26.24 |
| Energy contribution | -25.92 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15979322 120 - 23771897 AUGUCCAUAAAACUGGUUGCAAACGCACAUAAACCAUUUAAGUCGAAAAGAAAUCGGCGCGCUGGCAACUCUAACGAGAUAAAAACAACUCGCAGCCACUUUGAAUGGUAUUUUAUUUUG ....((((.(((.(((((((.............(((.....(((((.......)))))....)))...(((....))).............))))))).)))..))))............ ( -27.10) >DroSec_CAF1 8524 120 - 1 AUGUCCAUAAAACUGGUUACAAACGCACAUAAACCAUUUAAGCCGAAAAGAAAUCGGCGCGCUGGCAACUCUAACGAGAUAAAAACAACUCGCAGCCACUUUGAAUGGUAUUUUAUUUUG .((((((......)))..)))...........((((((((((((((.......)))))....((((..(((....))).......(.....)..))))..)))))))))........... ( -25.80) >DroSim_CAF1 9206 120 - 1 AUGUCCAUAAAACUGGUUGCAAACGCACAUAAACCAUUUAAGCCGAAAAGAAAUCGGCGCGCUGGCAACUCUAACGAGAUAAAAACAACUUGCAGCCACUUUGAAUGGUAUUUUAUUUUG ....((((.(((.(((((((((...........(((.....(((((.......)))))....)))...(((....)))...........))))))))).)))..))))............ ( -31.20) >DroEre_CAF1 8711 118 - 1 AUGUCCAUAAAACUGGUUGCAAACGCACAUAAACCAUUUAAGCCGAAAAGAAAUCGGCGCGCUGGCAACUCUAACGAGAUA--AACAACUCGGAGCCACUUUGAAUGGUAUUUUAUUUUG (((((((......)))..((....))))))..((((((((((((((.......)))))....((((..(((....)))...--..(.....)..))))..)))))))))........... ( -27.70) >DroYak_CAF1 8649 118 - 1 AUGUCCAUAAAACUGGUUGCAAACGCACAUAAACCAUUUAAGCCGAAAAGAAAUCGGCGCGCUGGCAACUCUAACGAGAUA--AACAACUCGGAGCCACUUUGAAUGGUAUUUUAUUUUG (((((((......)))..((....))))))..((((((((((((((.......)))))....((((..(((....)))...--..(.....)..))))..)))))))))........... ( -27.70) >consensus AUGUCCAUAAAACUGGUUGCAAACGCACAUAAACCAUUUAAGCCGAAAAGAAAUCGGCGCGCUGGCAACUCUAACGAGAUAAAAACAACUCGCAGCCACUUUGAAUGGUAUUUUAUUUUG .((((((......)))..)))...........((((((((((((((.......)))))....((((..(((....))).......(.....)..))))..)))))))))........... (-26.24 = -25.92 + -0.32)

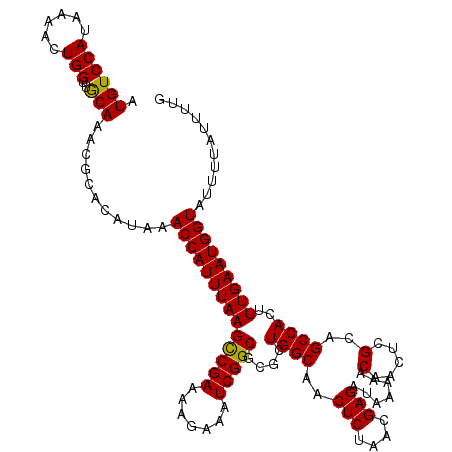

| Location | 15,979,362 – 15,979,478 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.80 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -33.72 |
| Energy contribution | -33.32 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15979362 116 - 23771897 UAU----AUAAUCAGGCCGCUCCGCCAGGCUGCGGAGGCGAUGUCCAUAAAACUGGUUGCAAACGCACAUAAACCAUUUAAGUCGAAAAGAAAUCGGCGCGCUGGCAACUCUAACGAGAU ...----........(((.((((((......))))))(((.............(((((((....))).....)))).....(((((.......))))).))).)))..(((....))).. ( -32.90) >DroSec_CAF1 8564 120 - 1 UAUACAUAUAAUCAGGCCGCUCCGCCAGGCUGCGGAGGCGAUGUCCAUAAAACUGGUUACAAACGCACAUAAACCAUUUAAGCCGAAAAGAAAUCGGCGCGCUGGCAACUCUAACGAGAU ...............(((.((((((......))))))(((.............(((((.............))))).....(((((.......))))).))).)))..(((....))).. ( -33.82) >DroSim_CAF1 9246 120 - 1 UAUACAUAUAAUCAGGCCGCUCCGCCAGGCUGCGGAGGCGAUGUCCAUAAAACUGGUUGCAAACGCACAUAAACCAUUUAAGCCGAAAAGAAAUCGGCGCGCUGGCAACUCUAACGAGAU ...............(((.((((((......))))))(((.............(((((((....))).....)))).....(((((.......))))).))).)))..(((....))).. ( -35.60) >DroEre_CAF1 8749 116 - 1 UAU----AUAAUCAGGCCGCUCCGUCAGGCUGCGGAGGCGAUGUCCAUAAAACUGGUUGCAAACGCACAUAAACCAUUUAAGCCGAAAAGAAAUCGGCGCGCUGGCAACUCUAACGAGAU ...----........(((.((((((......))))))(((.............(((((((....))).....)))).....(((((.......))))).))).)))..(((....))).. ( -33.90) >DroYak_CAF1 8687 116 - 1 UAU----AUAAUCAGGCCGCUCCGUCAGGCUGCGGAGGCGAUGUCCAUAAAACUGGUUGCAAACGCACAUAAACCAUUUAAGCCGAAAAGAAAUCGGCGCGCUGGCAACUCUAACGAGAU ...----........(((.((((((......))))))(((.............(((((((....))).....)))).....(((((.......))))).))).)))..(((....))).. ( -33.90) >consensus UAU____AUAAUCAGGCCGCUCCGCCAGGCUGCGGAGGCGAUGUCCAUAAAACUGGUUGCAAACGCACAUAAACCAUUUAAGCCGAAAAGAAAUCGGCGCGCUGGCAACUCUAACGAGAU ...............(((.((((((......))))))(((.............(((((.............))))).....(((((.......))))).))).)))..(((....))).. (-33.72 = -33.32 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:20 2006