
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,978,853 – 15,979,013 |
| Length | 160 |
| Max. P | 0.989462 |

| Location | 15,978,853 – 15,978,973 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.35 |
| Mean single sequence MFE | -22.71 |
| Consensus MFE | -18.54 |
| Energy contribution | -19.54 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
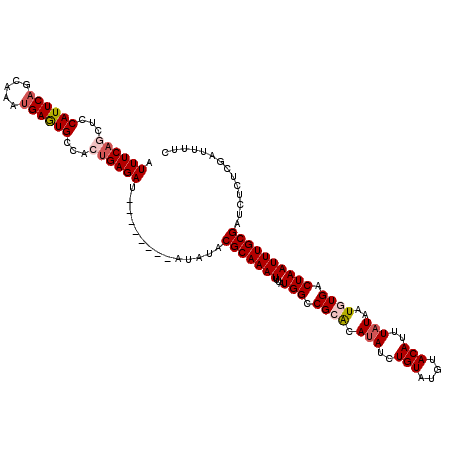
>3L_DroMel_CAF1 15978853 120 + 23771897 AUUUCAACUCCAUUCAGCAAAUGAGUGCCACAGAGAUACAUACAUAUAUACGCAAAUUUAUGGCCGCGCAUAUCUGUAUGUACACUUAUAAUGUGACUAAUUUGCGAUCUCUCGAUUUUC ................((((((.(((..(((((((.((((((((.((((.(((............))).)))).))))))))..)))....))))))).))))))((((....))))... ( -26.00) >DroSec_CAF1 8055 112 + 1 UUUUCAGCUCCAUUCAGCAAAUGAGUGCCACUGAGAU--------AUAUACGCAAAUUUAUGGCCGCACAUAUCUGUAUGUACAUUUAUAAUGUGACUAAUUUGCGAUCUCUCGAUUUUC .((((((...((((((.....))))))...)))))).--------.....(((((((...(((.((((.(((..(((....)))..)))..)))).)))))))))).............. ( -24.20) >DroSim_CAF1 8736 112 + 1 UUUUCAACUCCAUUCAGCAAAUGAGUGCCACUGAGAU--------AUAUACGCAAAUUUAUGGCCGCACAUAUCUGUAUGUACAUUUAUAAUGUGACUAAUUUGCGAUCUCUCGAUUUUC ...((.....((((((.....)))))).....(((((--------.....(((((((...(((.((((.(((..(((....)))..)))..)))).)))))))))))))))..))..... ( -21.30) >DroEre_CAF1 8290 110 + 1 AUUUCAGCUCCAUUCAGCAAAUGAAUGCCACUGAGAU----------AUACGCAAAUUUAUGGCCGCACAUAUCUGUAUGUACAUUUAUAAUCUGACUAAUUUGCGAUCUCUCGAUUUUC (((((((...((((((.....))))))...)))))))----------...((((((((((((......))))((..((((......))))....))..)))))))).............. ( -22.10) >DroYak_CAF1 8228 110 + 1 AUUUCAGCUCCAUUCAGCAAACGAAUGCCACUGAGAU----------AUACGCAAAUUUAUGGCCGCACACAUCUGUAUGUACAUUUAUAAUCUGACUAAUUUGCGAUCUCUCGAUUUUC (((((((...(((((.......)))))...)))))))----------...((((((((.(((..((.(((....))).))..))).............)))))))).............. ( -19.94) >consensus AUUUCAGCUCCAUUCAGCAAAUGAGUGCCACUGAGAU________AUAUACGCAAAUUUAUGGCCGCACAUAUCUGUAUGUACAUUUAUAAUGUGACUAAUUUGCGAUCUCUCGAUUUUC .((((((...((((((.....))))))...))))))..............(((((((...(((.((((.(((..(((....)))..)))..)))).)))))))))).............. (-18.54 = -19.54 + 1.00)

| Location | 15,978,893 – 15,979,013 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.86 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -25.34 |
| Energy contribution | -25.46 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15978893 120 - 23771897 AUAGAAGAUUGAAGCCCAGAUUGGAUGAGUCUGGGAUUUGGAAAAUCGAGAGAUCGCAAAUUAGUCACAUUAUAAGUGUACAUACAGAUAUGCGCGGCCAUAAAUUUGCGUAUAUAUGUA ......((((((..((((((((.....)))))))).((((((...((....)))).))))))))))((((.....))))...((((.((((((((((........)))))))))).)))) ( -33.40) >DroSec_CAF1 8092 115 - 1 AUAGAAGAUUGAAGCCCAGAUUGGAUGAGUCUGGGAUUUGGAAAAUCGAGAGAUCGCAAAUUAGUCACAUUAUAAAUGUACAUACAGAUAUGUGCGGCCAUAAAUUUGCGUAUAU----- ........(..((.((((((((.....)))))))).))..)...(((....)))((((((((.(((.(((.(((..(((....)))..)))))).)))....)))))))).....----- ( -29.20) >DroSim_CAF1 8773 115 - 1 AUAGAAGAUUGAAGCCCAGAUUGGAUGAGUCUGGGAUUUGGAAAAUCGAGAGAUCGCAAAUUAGUCACAUUAUAAAUGUACAUACAGAUAUGUGCGGCCAUAAAUUUGCGUAUAU----- ........(..((.((((((((.....)))))))).))..)...(((....)))((((((((.(((.(((.(((..(((....)))..)))))).)))....)))))))).....----- ( -29.20) >DroEre_CAF1 8327 113 - 1 AUAGAAGAUUGAAGCCCAGAUUGGAUGAGUCUUGGAUUUGGAAAAUCGAGAGAUCGCAAAUUAGUCAGAUUAUAAAUGUACAUACAGAUAUGUGCGGCCAUAAAUUUGCGUAU------- ...............(((((((.((......)).)))))))...(((....)))((((((((........(((...((((((((....))))))))...)))))))))))...------- ( -22.60) >DroYak_CAF1 8265 113 - 1 AUAGAAGAUUGAAGCCCGGAUUGGAUGAGUCUUGGAUUUGGAAAAUCGAGAGAUCGCAAAUUAGUCAGAUUAUAAAUGUACAUACAGAUGUGUGCGGCCAUAAAUUUGCGUAU------- ........(..((.((.(((((.....))))).)).))..)...(((....)))((((((((........(((...((((((((....))))))))...)))))))))))...------- ( -22.00) >consensus AUAGAAGAUUGAAGCCCAGAUUGGAUGAGUCUGGGAUUUGGAAAAUCGAGAGAUCGCAAAUUAGUCACAUUAUAAAUGUACAUACAGAUAUGUGCGGCCAUAAAUUUGCGUAUAU_____ ........(..((.((((((((.....)))))))).))..)...(((....)))((((((((.(((..((.....))(((((((....))))))))))....)))))))).......... (-25.34 = -25.46 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:17 2006