
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,972,302 – 15,972,443 |
| Length | 141 |
| Max. P | 0.793023 |

| Location | 15,972,302 – 15,972,418 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -40.39 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.58 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
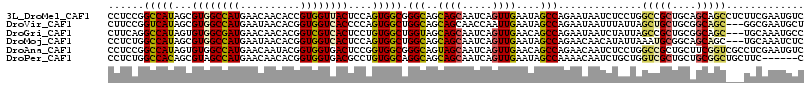
>3L_DroMel_CAF1 15972302 116 - 23771897 CCUCCGGCCAUAGCGUGGCCAUGAACAACACCGUGGUUACUCCAGUGGCGGGCAGCAGCAAUCAGUUGAAUAGCCAGAAUAAUCUCCUGGCCGCUGCAGCAGCCUCUUCGAAUGUC ....(((((((.(.(((((((((........))))))))).)..)))))((((.(((((..((....))...(((((.........))))).)))))....))))...))...... ( -39.90) >DroVir_CAF1 50658 113 - 1 CUUCCGGUCAUAGCGUGGCCAUGAAUAACACGGUGGUCACCCCAGUGGCUGGCAGCAGCAACCAAUUGAAUAGCCAGAAUAAUUUAUUAGCUGCUGCGGCAGC---GGCGAAUGCU ....(((((((.(.((((((((..........)))))))).)..)))))))((((((((......(((......)))((((...)))).))))))))((((..---......)))) ( -35.70) >DroGri_CAF1 53268 113 - 1 CUUCAGGCCAUAGUGUGGCGAUGAACAACACGGUCGUCACUCCUGUGGCUGGUAGCAGCAAUCAGUUGAACAGCCAGAAUAAUCUAUUAGCCGCUGCGGCAGC---UGCAAAUGCC .....((((((((.((((((((..........))))))))..))))))))(((((((((..((....))...((((((....)))....((....))))).))---)))...)))) ( -39.30) >DroMoj_CAF1 58009 113 - 1 CCUCUGGCCAUAGCGUGGCCAUGAAUAACACGGUGGUCACUCCAGUGGCUGGCAGCAGCAAUCAGUUGAAUAGCCAGAACAACAUAUUAAAUGCGGCAGCAGC---UGCAAAUCUC ((.(((((....))((((((((..........))))))))..))).))(((((..((((.....))))....)))))..............((((((....))---))))...... ( -36.40) >DroAna_CAF1 1681 116 - 1 CCUCCGGCCAUAGUGUGGCCAUGAACAAUACGGUGGUGACUCCGGUGGCGGGCAGUAGCAAUCAGUUGAACAGCCAGAACAAUCUCCUGGCCGCUGCUUCGGUCGCCUCGAAUGUC .....((((((...))))))..........(((.(((((((..((..((((.(((.((.......(((......)))......)).))).))))..))..))))))))))...... ( -43.02) >DroPer_CAF1 7443 110 - 1 CCUCUGGCCACAGCGUAGCCAUGAACAACACGGUGGUGACGCCUGUGGCAGGCAGCAGCAAUCAGUUGAAUAGCCAAAACAAUCUGCUGGUCGCUGCUGCGGCUGCUUC------C ...(((....))).((((((........(((((((....)))).)))....((((((((.((((((.((.............)).)))))).))))))))))))))...------. ( -48.02) >consensus CCUCCGGCCAUAGCGUGGCCAUGAACAACACGGUGGUCACUCCAGUGGCUGGCAGCAGCAAUCAGUUGAAUAGCCAGAACAAUCUACUAGCCGCUGCAGCAGC___UGCGAAUGUC ......(((((...((((((((..........))))))))....))))).(((..((((.....))))....)))..............(((((....)))))............. (-26.60 = -26.58 + -0.02)

| Location | 15,972,339 – 15,972,443 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -22.91 |
| Energy contribution | -22.55 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15972339 104 - 23771897 GCAGCAGCGGCAGCUUGUGCCAAUGCCUCCGGCCAUAGCGUGGCCAUGAACAACACCGUGGUUACUCCAGUGGCGGGCAGCAGCAAUCAGUUGAAUAGCCAGAA ((..((((((((.....))))..(((..((.(((((.(.(((((((((........))))))))).)..))))).))..))).......))))....))..... ( -37.50) >DroVir_CAF1 50692 95 - 1 -------C--CGCCAUGUGUAAACACUUCCGGUCAUAGCGUGGCCAUGAAUAACACGGUGGUCACCCCAGUGGCUGGCAGCAGCAACCAAUUGAAUAGCCAGAA -------(--((((..(((....)))....((((((...))))))...........)))))((((....))))(((((.....(((....)))....))))).. ( -26.20) >DroPse_CAF1 7456 103 - 1 -CAUCGGCCAUACCAUGCGUGAACACCUCUGGCCACAGCGUAGCCAUGAACAACACGGUGGUGACGCCCGUGGCAGGCAGCAGCAAUCAGUUGAAUAGCCAAAA -....(((....(((((((((..((....))..))).)))).((((((........((((....)))))))))).))...((((.....))))....))).... ( -31.90) >DroGri_CAF1 53302 95 - 1 -------C--CGUCCUGUGUCAACACUUCAGGCCAUAGUGUGGCGAUGAACAACACGGUCGUCACUCCUGUGGCUGGUAGCAGCAAUCAGUUGAACAGCCAGAA -------.--....((((.(((((......((((((((.((((((((..........))))))))..))))))))(((.......))).)))))))))...... ( -30.80) >DroMoj_CAF1 58043 95 - 1 -------C--CGGCGUGCGUGAACACCUCUGGCCAUAGCGUGGCCAUGAAUAACACGGUGGUCACUCCAGUGGCUGGCAGCAGCAAUCAGUUGAAUAGCCAGAA -------(--((.((((.(....)...(((((((((...))))))).))....)))).)))((((....))))(((((..((((.....))))....))))).. ( -33.10) >DroPer_CAF1 7474 103 - 1 -CAUCGGCCAUACCAUGCGUGAACACCUCUGGCCACAGCGUAGCCAUGAACAACACGGUGGUGACGCCUGUGGCAGGCAGCAGCAAUCAGUUGAAUAGCCAAAA -....(((.......((((....)....(((.(((((((((.(((((..........))))).))).)))))))))))).((((.....))))....))).... ( -33.10) >consensus _______C__CACCAUGCGUGAACACCUCUGGCCAUAGCGUGGCCAUGAACAACACGGUGGUCACUCCAGUGGCUGGCAGCAGCAAUCAGUUGAAUAGCCAGAA ...............................(((((...((((((((..........))))))))....))))).(((..((((.....))))....))).... (-22.91 = -22.55 + -0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:11 2006