
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,969,731 – 15,969,824 |
| Length | 93 |
| Max. P | 0.802156 |

| Location | 15,969,731 – 15,969,824 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -16.61 |
| Energy contribution | -16.92 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
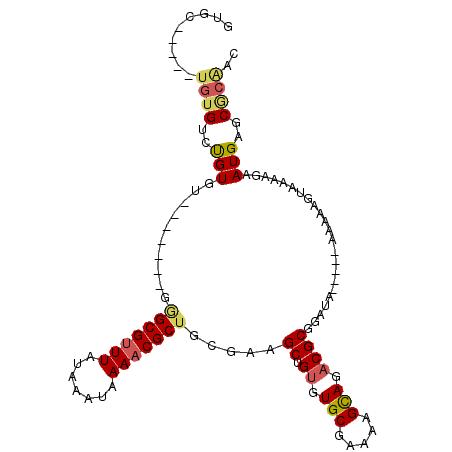
>3L_DroMel_CAF1 15969731 93 - 23771897 GUGC-----UGUGUCUGUGU-------GGGCGUUUAUAAAUAAAACGCUGCGAAGCUGUGUGCGAAAAGCAGACGCGGAUA-----AAGAAGUAAAAGAAUGAGCGCAAC ((((-----(.(((((((((-------.(((((((.......)))))))((...((.....)).....))..)))))))))-----................)))))... ( -26.03) >DroVir_CAF1 49754 91 - 1 --UG-----UUUGUGUGUGUCGAGUGAGUGCG-UUAUAAAUAAAACGCCAAGAAGCUGCGUGCAAAAAGUAGACGCCAAGA-----AAAAUAA---AGAAUGUGCCC--- --((-----(((.((.(((((......(.(((-((........)))))).....((.....))........)))))))...-----.))))).---...........--- ( -16.30) >DroSec_CAF1 72313 93 - 1 GUGC-----UGUGUCUGUGU-------GGGCGUUUAUAAAUAAAACGCUGCGAAGCUGUGUGCGAAAAGUAGACGCGGAUA-----AAGAAGUAAAAGAAUGAGCGCAAC ((((-----(.(((((((((-------.(((((((.......))))))).....((.....)).........)))))))))-----................)))))... ( -23.93) >DroSim_CAF1 71547 93 - 1 GUGC-----UGUGUCUGUGU-------GGGCGUUUAUAAAUAAAACGCUGCGAAGCUGUGUGCGAAAAGCAGACGCGGAUA-----AAGAAGUAAAAGAAUGAGCGCAAC ((((-----(.(((((((((-------.(((((((.......)))))))((...((.....)).....))..)))))))))-----................)))))... ( -26.03) >DroEre_CAF1 68925 93 - 1 GUGU-----UGUGUCUGUGU-------GAGCGUUUAUAAAUAAAACGCUGCGAAGCUGUGUGCGAAAAGCAGACGCGGAUA-----AAAAAGUAAAAGAAUGAGCGCAAC ..((-----(((((..((((-------.(((((((.......)))))))))...((.((.(((.....))).)))).....-----.............))..))))))) ( -24.60) >DroAna_CAF1 39781 103 - 1 GUGGUCGAGUGUGUGCGUGU-------GGGCGUUUAUAAAUAAAACGCUGCGAAGCUGUGUGCAAAUAGCAGACGCGGAUAGAUGGAAAAAAUAAAAGAAUGAGCACGAC (((.(((.((.(((.(((((-------.(((((((.......))))))).....(((((......)))))..))))).))).))................))).)))... ( -27.03) >consensus GUGC_____UGUGUCUGUGU_______GGGCGUUUAUAAAUAAAACGCUGCGAAGCUGUGUGCGAAAAGCAGACGCGGAUA_____AAAAAGUAAAAGAAUGAGCGCAAC .........((((..(((..........(((((((.......))))))).....((.((.(((.....))).)))).......................)))..)))).. (-16.61 = -16.92 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:08 2006