| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,946,760 – 15,946,869 |
| Length | 109 |
| Max. P | 0.890611 |

| Location | 15,946,760 – 15,946,869 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -23.31 |
| Energy contribution | -24.48 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
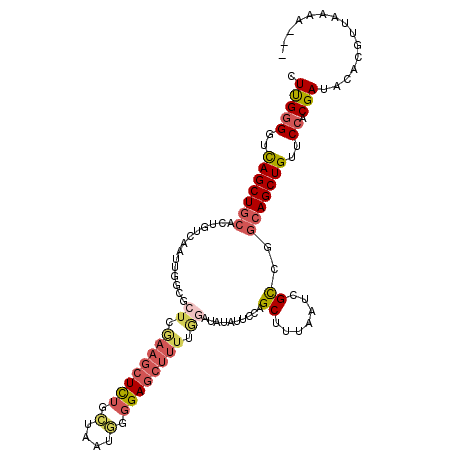
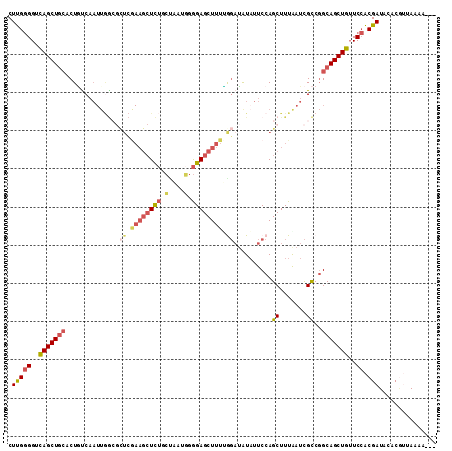
>3L_DroMel_CAF1 15946760 109 + 23771897 CUUGGGGUCAGCUGCACUGCCAAUUGGCGCUCGAAGCUCUGCUAAUGGGGAGCUUUCGGAUAUAUUCCAGCUUUAAUCGCCGGCAGCUGUUCCACGAUACACGUCAAAA--- ..(((((.(((((((...(((....)))(((.((((((((.(....).)))))))).(((.....))))))...........)))))))))))).(((....)))....--- ( -40.80) >DroSec_CAF1 48382 109 + 1 CUUGGGGUCAGCUGCACUGUCAAUUGGCGCUCGAAGCUCUGCUAAUUGGGAGCUUUUGGAUGUAUUCCAGCUUUAAUCGCCGGCAGCUGUUCCACGAUACACGUUAAAA--- ..(((((.(((((((.((.(((((((((((.....))...))))))))).))...(((((.....)))))............))))))))))))...............--- ( -35.40) >DroSim_CAF1 48473 109 + 1 CUUGGGGUCAGCUGCACUAUCAAUUGGCGCUCGAAGCUCUGCCAAUGGGGAGCUUUUGGAUGUAUUCCAGCUUUAAUCGCCUGCAGCUGUUCCACGAUACACGUUAAAA--- ..(((((.((((((((.........((((...((((((((.(....).))))))))((((.....))))........)))))))))))))))))...............--- ( -37.50) >DroEre_CAF1 45776 93 + 1 CUUGGGGGCAGCUGAACUGUC----------------UCUGCGAAUGGGGAGCUUUCAGAUAUAUUCCAGCUUUGAUCGCCGGCAGCUGUUCCACGAUACACGUUAAAA--- .(((((((((((((..(((.(----------------(((.(....).))))....))).......((.((.......)).)))))))))))).)))............--- ( -27.30) >DroYak_CAF1 55091 108 + 1 CUUGGGGUCAGCUGCACUGUCAAUUGGCGCUCGAAGCUCUUCUAAUGGGGAGCUUUUAGAUAUAUUCCAGCUUUGAUCGCCGGCAGCUGUUCCACGAUACACC-UAAAA--- ..(((((.(((((((.........(((((.((((((((..(((((.((....)).)))))........)))))))).))))))))))))))))).........-.....--- ( -37.40) >DroAna_CAF1 18652 110 + 1 CUCGAGGUUAGCUCCACUCUCAAUUAA--UUGAAAGCUUCAGCCUCCAAGAGCUUUGGCCGAUCUUCCAGCUUUAAUUGUCGGCAGCUGGUCGGCGAUAAACAAAAAAAAAA (((((((((((((......((((....--)))).))))..))))))...))).....(((((....((((((............)))))))))))................. ( -25.90) >consensus CUUGGGGUCAGCUGCACUGUCAAUUGGCGCUCGAAGCUCUGCUAAUGGGGAGCUUUUGGAUAUAUUCCAGCUUUAAUCGCCGGCAGCUGUUCCACGAUACACGUUAAAA___ .(((((..(((((((..............((.((((((((.(....).)))))))).))..........((.......))..)))))))..)).)))............... (-23.31 = -24.48 + 1.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:02 2006