
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,934,047 – 15,934,137 |
| Length | 90 |
| Max. P | 0.939523 |

| Location | 15,934,047 – 15,934,137 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 86.92 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -23.09 |
| Energy contribution | -22.40 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
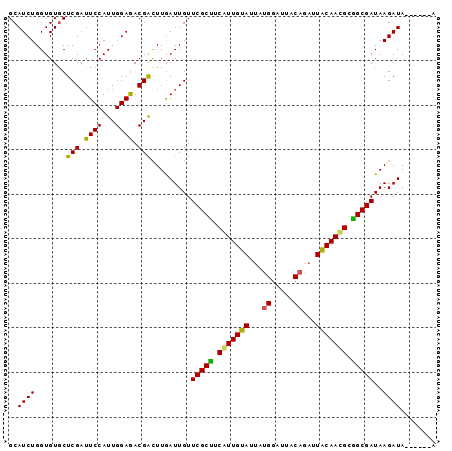
>3L_DroMel_CAF1 15934047 90 + 23771897 GCAUCUGGUGUGCUCGUUUCCAUUGGAGACGACUUGAUUGUUCGCUUCGUUGUAUUAUGGAUUACAGAUUACAACGCGGCGAUAAGAUA------A ((((.....))))((((((((...))))))))....(((..(((((.(((((((...((.....))...))))))).)))))...))).------. ( -30.20) >DroSec_CAF1 35856 90 + 1 GCAUCUGGUGUGCUCGAUUCCAUUGGAGACGGCUUGAUUGUUCGCUUCGUUGUAUUAUGGAUUACAGAUUACAACGCGGCGAUAAGAUA------A ..((((.....((.((.((((...)))).))))........(((((.(((((((...((.....))...))))))).)))))..)))).------. ( -25.90) >DroEre_CAF1 33445 90 + 1 GCAUCUGGUGUACUCGAUUCCAUUGGAGACGACUUGAUUGUUCGCCUCAUUGUAUUAUGGAUUACAGAUUGCAACGCGGCGAUAAGAUA------A (((((.(((((.(((.(......).))))).))).)).)))(((((.(.(((((...((.....))...))))).).))))).......------. ( -21.90) >DroAna_CAF1 440 93 + 1 GCAUCUGGUGUGCUCGAUUCCAUUGGAAACGAUUUAAUUGUUCGCUCCAUUGUA---CGGAUUACAGAUUACAAUGAAGCGAUAAGAUAUGAGAAG ...(((.((((.(((((((..((((....))))..))))).(((((.(((((((---............))))))).)))))..)))))).))).. ( -24.50) >consensus GCAUCUGGUGUGCUCGAUUCCAUUGGAGACGACUUGAUUGUUCGCUUCAUUGUAUUAUGGAUUACAGAUUACAACGCGGCGAUAAGAUA______A ..((((.......(((.((((...)))).))).........(((((.(((((((...((.....))...))))))).)))))..))))........ (-23.09 = -22.40 + -0.69)


| Location | 15,934,047 – 15,934,137 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 86.92 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -17.75 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
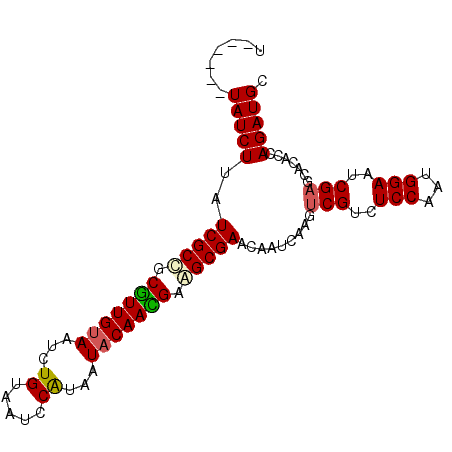
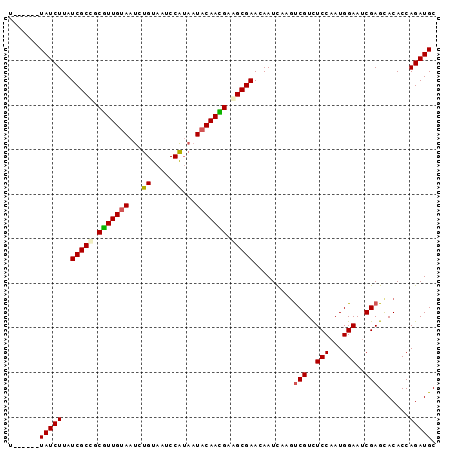
>3L_DroMel_CAF1 15934047 90 - 23771897 U------UAUCUUAUCGCCGCGUUGUAAUCUGUAAUCCAUAAUACAACGAAGCGAACAAUCAAGUCGUCUCCAAUGGAAACGAGCACACCAGAUGC .------(((((..((((..(((((((.(.((.....)).).)))))))..)))).........((((.(((...))).)))).......))))). ( -21.20) >DroSec_CAF1 35856 90 - 1 U------UAUCUUAUCGCCGCGUUGUAAUCUGUAAUCCAUAAUACAACGAAGCGAACAAUCAAGCCGUCUCCAAUGGAAUCGAGCACACCAGAUGC .------(((((..((((..(((((((.(.((.....)).).)))))))..))))........((((..(((...)))..)).)).....))))). ( -21.60) >DroEre_CAF1 33445 90 - 1 U------UAUCUUAUCGCCGCGUUGCAAUCUGUAAUCCAUAAUACAAUGAGGCGAACAAUCAAGUCGUCUCCAAUGGAAUCGAGUACACCAGAUGC .------(((((..(((((.(((((.....((.....)).....))))).))))).......(.(((..(((...)))..))).).....))))). ( -19.10) >DroAna_CAF1 440 93 - 1 CUUCUCAUAUCUUAUCGCUUCAUUGUAAUCUGUAAUCCG---UACAAUGGAGCGAACAAUUAAAUCGUUUCCAAUGGAAUCGAGCACACCAGAUGC .......(((((..(((((((((((((............---))))))))))))).........(((.((((...)))).))).......))))). ( -24.50) >consensus U______UAUCUUAUCGCCGCGUUGUAAUCUGUAAUCCAUAAUACAACGAAGCGAACAAUCAAGUCGUCUCCAAUGGAAUCGAGCACACCAGAUGC .......(((((..(((((.(((((((...((.....))...))))))).))))).........(((..(((...)))..))).......))))). (-17.75 = -18.00 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:58 2006