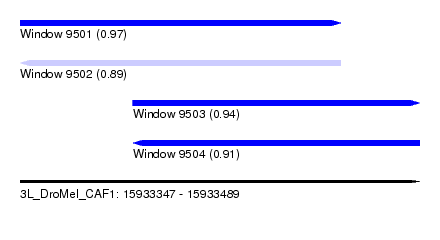
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,933,347 – 15,933,489 |
| Length | 142 |
| Max. P | 0.969256 |

| Location | 15,933,347 – 15,933,461 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.28 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -28.46 |
| Energy contribution | -28.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15933347 114 + 23771897 AUAAUUCUCAUAAUUGAGAUAUAUCUAGUAUAUUUUAAGUUCCGAACUGCGGC-UUCGGUCACACUGGCGGCGACGUCGGCAGCGAUGCCGACGUCUAUUUACAACGCAUCGUUU .............(((((((((((...)))))))))))....(((..((((((-(((((.....)))).)))((((((((((....)))))))))).........)))))))... ( -37.70) >DroSec_CAF1 35155 115 + 1 AUAAUUCCCAUAAUUGAGAUAUUUCUAGUAUAUUUUAUGUUCCGAACUGUGGCCUUCGGUCACACUGGUUGCGACGUCGGCAGCGAUGCCGACGUCUAUUUACAACGCAUCGUUU ..............((((((((.......)))))))).((.(((...(((((((...))))))).)))..))((((((((((....))))))))))................... ( -33.50) >DroSim_CAF1 35418 115 + 1 AUAAUUCCCAUAAUUGAGAUAUUUCUAGUAUAUUUUAAGUUCCGAACUGCGGCCUUCGGUCACACUGGCUGCGACGUCGGCAGCGAUGCCGACGUCUAUUUACAACGCAUCGUUU .............(((((((((.......)))))))))....(((...((((((...(....)...))))))((((((((((....)))))))))).............)))... ( -36.40) >DroEre_CAF1 32749 114 + 1 AUCAUUAAUAGAAUUGAGGUUUUUCUAGUAUAUUUAAA-CUCUGAAUUGUCCCCCUUGGUCACACUGGCAGCGACGUCGGCAGAGAUGCCGACGUCUAUUUACAACGCAUCGUUU ........(((((.........)))))........(((-(..((..((((........(((.....)))...((((((((((....)))))))))).....))))..))..)))) ( -27.60) >consensus AUAAUUCCCAUAAUUGAGAUAUUUCUAGUAUAUUUUAAGUUCCGAACUGCGGCCUUCGGUCACACUGGCUGCGACGUCGGCAGCGAUGCCGACGUCUAUUUACAACGCAUCGUUU .............(((((((((.......)))))))))....(((..((((((..((((.....))))..))((((((((((....)))))))))).........)))))))... (-28.46 = -28.90 + 0.44)


| Location | 15,933,347 – 15,933,461 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.28 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -26.45 |
| Energy contribution | -27.95 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15933347 114 - 23771897 AAACGAUGCGUUGUAAAUAGACGUCGGCAUCGCUGCCGACGUCGCCGCCAGUGUGACCGAA-GCCGCAGUUCGGAACUUAAAAUAUACUAGAUAUAUCUCAAUUAUGAGAAUUAU ...(((((((.........((((((((((....))))))))))..(((....)))......-..))))..))).......................(((((....)))))..... ( -31.20) >DroSec_CAF1 35155 115 - 1 AAACGAUGCGUUGUAAAUAGACGUCGGCAUCGCUGCCGACGUCGCAACCAGUGUGACCGAAGGCCACAGUUCGGAACAUAAAAUAUACUAGAAAUAUCUCAAUUAUGGGAAUUAU ......((.(((((.....((((((((((....)))))))))))))))))((((..(((((........))))).)))).................(((((....)))))..... ( -34.30) >DroSim_CAF1 35418 115 - 1 AAACGAUGCGUUGUAAAUAGACGUCGGCAUCGCUGCCGACGUCGCAGCCAGUGUGACCGAAGGCCGCAGUUCGGAACUUAAAAUAUACUAGAAAUAUCUCAAUUAUGGGAAUUAU ...(((.((..........((((((((((....))))))))))((.(((..((....))..))).)).))))).......................(((((....)))))..... ( -33.70) >DroEre_CAF1 32749 114 - 1 AAACGAUGCGUUGUAAAUAGACGUCGGCAUCUCUGCCGACGUCGCUGCCAGUGUGACCAAGGGGGACAAUUCAGAG-UUUAAAUAUACUAGAAAAACCUCAAUUCUAUUAAUGAU ((((..((.(((((.....((((((((((....)))))))))).((.((..((....)).)).))))))).))..)-)))........(((((.........)))))........ ( -31.00) >consensus AAACGAUGCGUUGUAAAUAGACGUCGGCAUCGCUGCCGACGUCGCAGCCAGUGUGACCGAAGGCCACAGUUCGGAACUUAAAAUAUACUAGAAAUAUCUCAAUUAUGGGAAUUAU ..(((.((.(((((.....((((((((((....))))))))))))))))).)))..(((((........)))))......................(((((....)))))..... (-26.45 = -27.95 + 1.50)

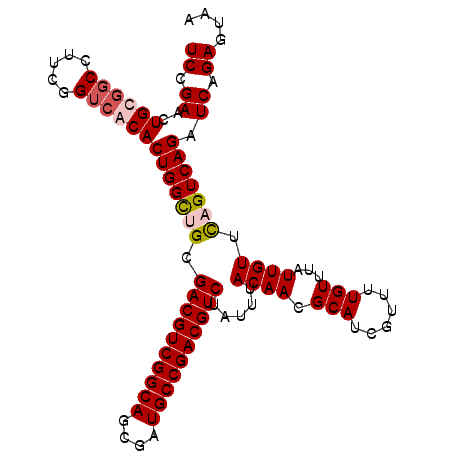
| Location | 15,933,387 – 15,933,489 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -30.40 |
| Energy contribution | -31.53 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15933387 102 + 23771897 UCCGAACUGCGGC-UUCGGUCACACUGGCGGCGACGUCGGCAGCGAUGCCGACGUCUAUUUACAACGCAUCGUUUUGUUUAUUGUUCAGUCAGAUCAGAGUAA ......(((.(((-....)))...(((((...((((((((((....)))))))))).....((((.(((......)))...))))...)))))..)))..... ( -35.00) >DroSec_CAF1 35195 103 + 1 UCCGAACUGUGGCCUUCGGUCACACUGGUUGCGACGUCGGCAGCGAUGCCGACGUCUAUUUACAACGCAUCGUUUUGUUUAUUGUUCAGUCAGAUCAGAGUAA ((.((..(((((((...)))))))(((..((.((((((((((....)))))))))).....((((.(((......)))...)))).))..))).)).)).... ( -35.70) >DroSim_CAF1 35458 103 + 1 UCCGAACUGCGGCCUUCGGUCACACUGGCUGCGACGUCGGCAGCGAUGCCGACGUCUAUUUACAACGCAUCGUUUUGUUUAUUGUUCAGUCAGAUCAGAGUAA ......(((.((((...))))...(((((((.((((((((((....)))))))))).....((((.(((......)))...)))).)))))))..)))..... ( -36.20) >DroEre_CAF1 32788 103 + 1 UCUGAAUUGUCCCCCUUGGUCACACUGGCAGCGACGUCGGCAGAGAUGCCGACGUCUAUUUACAACGCAUCGUUUUGUUUAUUGUUUAGUCAGAUCAGAGUAA (((((..((.((.....)).))..(((((...((((((((((....)))))))))).....((((.(((......)))...))))...))))).))))).... ( -34.90) >consensus UCCGAACUGCGGCCUUCGGUCACACUGGCUGCGACGUCGGCAGCGAUGCCGACGUCUAUUUACAACGCAUCGUUUUGUUUAUUGUUCAGUCAGAUCAGAGUAA ((.((..((((((.....))))))(((((((.((((((((((....)))))))))).....((((.(((......)))...)))).))))))).)).)).... (-30.40 = -31.53 + 1.13)


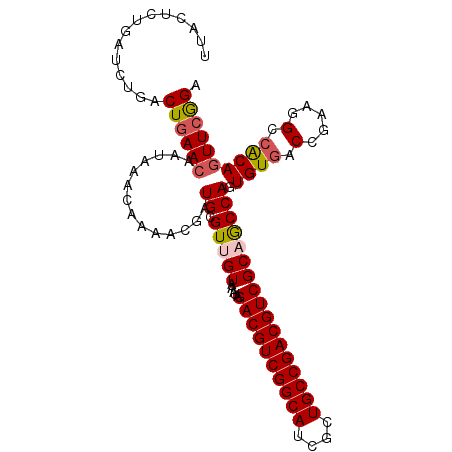
| Location | 15,933,387 – 15,933,489 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -29.12 |
| Energy contribution | -29.75 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15933387 102 - 23771897 UUACUCUGAUCUGACUGAACAAUAAACAAAACGAUGCGUUGUAAAUAGACGUCGGCAUCGCUGCCGACGUCGCCGCCAGUGUGACCGAA-GCCGCAGUUCGGA ((((.(((..........(((((...((......)).))))).....((((((((((....)))))))))).....))).))))(((((-.......))))). ( -30.80) >DroSec_CAF1 35195 103 - 1 UUACUCUGAUCUGACUGAACAAUAAACAAAACGAUGCGUUGUAAAUAGACGUCGGCAUCGCUGCCGACGUCGCAACCAGUGUGACCGAAGGCCACAGUUCGGA ..............((((((..............((.(((((.....((((((((((....))))))))))))))))).((((.((...)).)))))))))). ( -34.90) >DroSim_CAF1 35458 103 - 1 UUACUCUGAUCUGACUGAACAAUAAACAAAACGAUGCGUUGUAAAUAGACGUCGGCAUCGCUGCCGACGUCGCAGCCAGUGUGACCGAAGGCCGCAGUUCGGA ..............((((((..........((((....)))).....((((((((((....))))))))))((.(((..((....))..))).)).)))))). ( -35.00) >DroEre_CAF1 32788 103 - 1 UUACUCUGAUCUGACUAAACAAUAAACAAAACGAUGCGUUGUAAAUAGACGUCGGCAUCUCUGCCGACGUCGCUGCCAGUGUGACCAAGGGGGACAAUUCAGA ....(((((.(((.....(((((...((......)).))))).....((((((((((....)))))))))).....)))(((..((...))..)))..))))) ( -29.00) >consensus UUACUCUGAUCUGACUGAACAAUAAACAAAACGAUGCGUUGUAAAUAGACGUCGGCAUCGCUGCCGACGUCGCAGCCAGUGUGACCGAAGGCCACAGUUCGGA ..............((((((..............((.(((((.....((((((((((....))))))))))))))))).((((.(.....).)))))))))). (-29.12 = -29.75 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:57 2006