| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,932,928 – 15,933,046 |
| Length | 118 |
| Max. P | 0.627217 |

| Location | 15,932,928 – 15,933,046 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -20.60 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
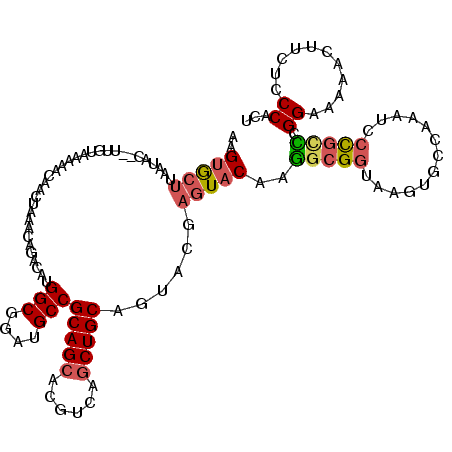
>3L_DroMel_CAF1 15932928 118 - 23771897 AAGUGCUUAAUAC--UUAUAAAAACAACUAAACAGACAUGGCGGAUGCCGCAGCACGUCAGCUGCAAUACGAGUACAAGGCGGUAAGUGUCAAAUCCCGCCCGGGAAAACUUCUCCCACU ..((((((.....--........................(((....)))(((((......))))).....))))))..(((((.............))))).((((.......))))... ( -33.12) >DroSec_CAF1 34726 112 - 1 AAGUGCU--AUAC---UGUAAAA--CACUAAACAGACAUGGCG-AUGCCGCAGCACGUCAGCUGCAGUACGAGUACAAGGCGGUAAGUGCCAAAUCCCGCCCGGGAAAACUUUUCCCACU ..(((((--.(((---((((...--..((....))...(((((-.(((....))))))))..)))))))..)))))..(((((.............))))).(((((.....)))))... ( -34.62) >DroSim_CAF1 34978 118 - 1 AAGUGCUUAAUAC--UUGUAAAAACAACUAAACAGACAUGGCGGAUGCCGCAGCACGUCAGCUGCAGUACGAGUACAAGGCGGUAAGUGCCAAAUCCCGUCCGGGAAAACUUUUCCCACU ....((((..(((--(((((...................(((....)))(((((......)))))..))))))))..))))(((....)))...........(((((.....)))))... ( -35.30) >DroEre_CAF1 32347 117 - 1 AAGUGGUAAAUAC--UCGUGAAAUCAACUAAACAGUCAUGGCGGAUGCCGCAGCACGUCAGCUGCAGUACGAGUACAAGGCGGUAAGUGCUAAAUACCGCCCGGAA-AACUUCUCCCACU .(((((....(((--(((((...................(((....)))(((((......)))))..))))))))(..(((((((.........)))))))..)..-........))))) ( -39.80) >DroYak_CAF1 31916 118 - 1 AAGUGCAAAAUAC--UUUUGAAAACAACUAAACAGACAUGGCGGAUGCCGCAGCACGUCAGCUGCAGUACGAGUACAAGGCGGUAAGUGUUAAAUCCCGCUCGGAAAAACUUUUCCCAUC ..((.((((....--.))))...))..............(((((..((((((((......))))).(((....)))..)))(((.........)))))))).(((((....))))).... ( -26.80) >DroMoj_CAF1 12067 118 - 1 CGGCACUUAAAACGAUAACAACU--AGCUAAACAAAUAUGGCGGAUGCCGCAGCACGUCAACUGCAGUACGAGUACAAAGCGGUAAGUAUUGAAAAAUAAUAGGAAGAAAAGCGGCCGCC .(((...................--.((((........))))....(((((.........(((((.(((....)))...)))))...((((....))))............))))).))) ( -25.50) >consensus AAGUGCUUAAUAC__UUGUAAAAACAACUAAACAGACAUGGCGGAUGCCGCAGCACGUCAGCUGCAGUACGAGUACAAGGCGGUAAGUGCCAAAUCCCGCCCGGAAAAACUUCUCCCACU ..(((((................................(((....)))(((((......)))))......)))))..(((((.............))))).((...........))... (-20.60 = -21.02 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:53 2006