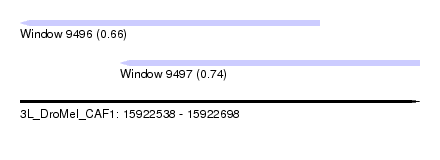
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,922,538 – 15,922,698 |
| Length | 160 |
| Max. P | 0.737708 |

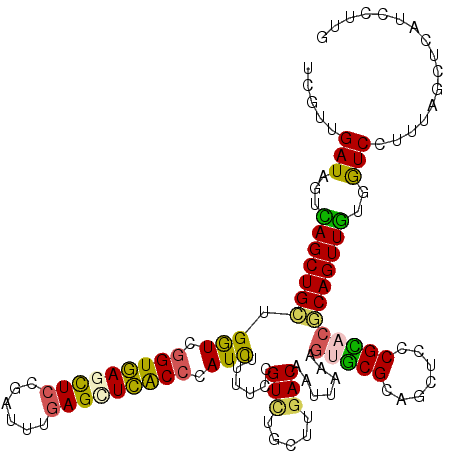
| Location | 15,922,538 – 15,922,658 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -30.75 |
| Energy contribution | -29.53 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15922538 120 - 23771897 UCGUUGAUAGUCAGCUGCUGGUCGGUGAGCUCCGAUUUGAGCUCACCCAUUUUUUCCGUCUGCUUGACAAUUAAAGUGCGCAGCUCCCGCACGCAGUUGUGGUCCUUUAGCUCAUCCUUG ..(((((..(.(((((((.....(((((((((......)))))))))..........(((.....))).......(((((.......))))))))))))....)..)))))......... ( -39.80) >DroVir_CAF1 3903 120 - 1 UCAUUGAUGGUUAGCUGUUGAUCAGUUAUUUCUGUCUUGAGUUGGCCAAUUUGUUCAGUUUGUUUGACAAUUAACGCGCGCAGCUCUCGUACACAGUUGUGAUCCUUAAGCUCGUCCUUG .....(((((.((((((.....))))))...)))))..(((((.((....((((.(((.....))))))).....)).(((((((.........))))))).......)))))....... ( -24.90) >DroSim_CAF1 24580 118 - 1 UCGUUGAUAGUCAGCUGCUGGUCGGUGAGCUCCGAU-UGAGCUCACCCAUCUUUUCCGUCUGCUUGACAAU-AAAGUGCGCAGCUCCCGCACGCAGUUGUGGUCCUUUAGCUCAUCCUUG ..(((((..(.(((((((.(((.(((((((((....-.))))))))).)))......(((.....)))...-...(((((.......))))))))))))....)..)))))......... ( -41.50) >DroEre_CAF1 21919 120 - 1 UCGUUGAUAGUCAGCUGCUGGUCGGUGAGCUCCGAUUUGAGCUCACCCAUCUUUUCCGUCUGCUUGACAAUUAAAGUGCGCAACUCCCGCACGCAGUUGUGGUCCUUUAGCUCAUCCUUG ..(((((..(.(((((((.(((.(((((((((......))))))))).)))......(((.....))).......(((((.......))))))))))))....)..)))))......... ( -41.70) >DroYak_CAF1 22525 120 - 1 UCGUUGAUUGUCAGCUGCUGGUCGGUGAGCUCCGAUUUGAGUUCACCCAUCUUUUCUGUCUGCUUGACAAUUAAAGUGCGCAGCUCCCGCACGCAGUUGUGGUCCUUUAGCUCAUCCUUG ..(((((..(.(((((((.(((.(((((((((......))))))))).))).....((((.....))))......(((((.......))))))))))))....)..)))))......... ( -39.60) >DroMoj_CAF1 2301 120 - 1 UCAUUGAUAGUUAGCUGCUGAUCGGUUAUUUCCGUUUUCAGUUGGCCUAUUUGCUCAGUUUGCUUCACAAUCAACACACGCAGCUCACGUACACAGUUAUGAUCCUUGAGCUCGUCCUUG ...(((((.((.(((.(((((.(((......)))...)))))((((......)).))....)))..)).)))))...(((.((((((((((......)))).....)))))))))..... ( -26.00) >consensus UCGUUGAUAGUCAGCUGCUGGUCGGUGAGCUCCGAUUUGAGCUCACCCAUCUUUUCCGUCUGCUUGACAAUUAAAGUGCGCAGCUCCCGCACGCAGUUGUGGUCCUUUAGCUCAUCCUUG .....(((...(((((((.(((.(((((((((......))))))))).)))......(((.....))).......(((((.......))))))))))))..)))................ (-30.75 = -29.53 + -1.21)


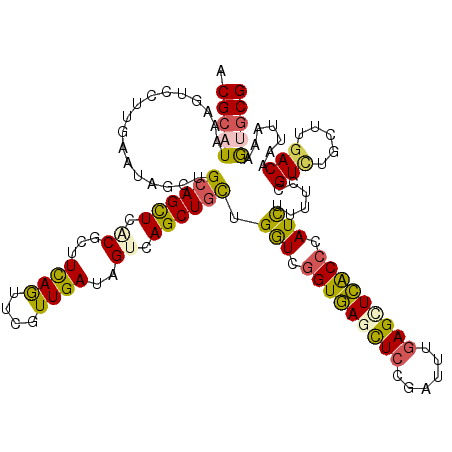
| Location | 15,922,578 – 15,922,698 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.10 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15922578 120 - 23771897 ACGCAUAAAGUCCUUGAAUAGCUGCAGCUCGCGCUUCAGUUCGUUGAUAGUCAGCUGCUGGUCGGUGAGCUCCGAUUUGAGCUCACCCAUUUUUUCCGUCUGCUUGACAAUUAAAGUGCG .(((((.........(((..(((((((((.((...((((....))))..)).)))))).))).(((((((((......)))))))))......))).(((.....))).......))))) ( -39.70) >DroVir_CAF1 3943 120 - 1 ACGCAUAAAAUCUUUGAAUAAUUGCAGUUCACGCUUUAGCUCAUUGAUGGUUAGCUGUUGAUCAGUUAUUUCUGUCUUGAGUUGGCCAAUUUGUUCAGUUUGUUUGACAAUUAACGCGCG .(((..........(((((((..(((((....))).(((((((..(((((.((((((.....))))))...))))).)))))))))....)))))))(((.....))).......))).. ( -27.70) >DroSim_CAF1 24620 117 - 1 ACGCAUAAAGUCC-UGAAUAGCUGAAGCUCGCGCUUCAGUUCGUUGAUAGUCAGCUGCUGGUCGGUGAGCUCCGAU-UGAGCUCACCCAUCUUUUCCGUCUGCUUGACAAU-AAAGUGCG .(((((...(.((-.....(((((((((....))))))))).((((.....))))....)).)(((((((((....-.)))))))))..........(((.....)))...-...))))) ( -42.00) >DroEre_CAF1 21959 120 - 1 ACGCAUAAAGUCCUUGAAUAGCUGCAGCUCUCGCUUCAGUUCGUUGAUAGUCAGCUGCUGGUCGGUGAGCUCCGAUUUGAGCUCACCCAUCUUUUCCGUCUGCUUGACAAUUAAAGUGCG .(((((...(((..(((...((((.(((....))).)))))))..))).((((((.(.(((..(((((((((......)))))))))........))).).)).)))).......))))) ( -39.60) >DroYak_CAF1 22565 120 - 1 ACGCAUAAAGUCCUUGAAUAGCUGCAGCUCACGCUUCAGUUCGUUGAUUGUCAGCUGCUGGUCGGUGAGCUCCGAUUUGAGUUCACCCAUCUUUUCUGUCUGCUUGACAAUUAAAGUGCG .(((((.............(((((.(((....))).)))))..((((((((((((.((.(((.(((((((((......))))))))).)))......))..)).)))))))))).))))) ( -41.10) >DroMoj_CAF1 2341 120 - 1 ACGCAUAAAAUCUUUAAAUAGCUGCAGUUCUCGCUUCAACUCAUUGAUAGUUAGCUGCUGAUCGGUUAUUUCCGUUUUCAGUUGGCCUAUUUGCUCAGUUUGCUUCACAAUCAACACACG ..(((.........(((((((((((((((..(...((((....))))..)..))))))(((.(((......)))...)))...)).))))))).......)))................. ( -21.59) >consensus ACGCAUAAAGUCCUUGAAUAGCUGCAGCUCACGCUUCAGUUCGUUGAUAGUCAGCUGCUGGUCGGUGAGCUCCGAUUUGAGCUCACCCAUCUUUUCCGUCUGCUUGACAAUUAAAGUGCG .(((((.................((((((.((...((((....))))..)).)))))).(((.(((((((((......))))))))).)))......(((.....))).......))))) (-28.70 = -28.10 + -0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:50 2006