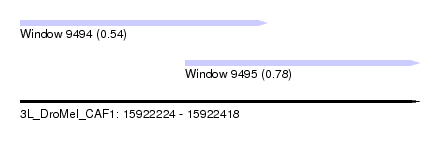
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,922,224 – 15,922,418 |
| Length | 194 |
| Max. P | 0.781498 |

| Location | 15,922,224 – 15,922,344 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -35.66 |
| Consensus MFE | -28.21 |
| Energy contribution | -28.41 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.19 |
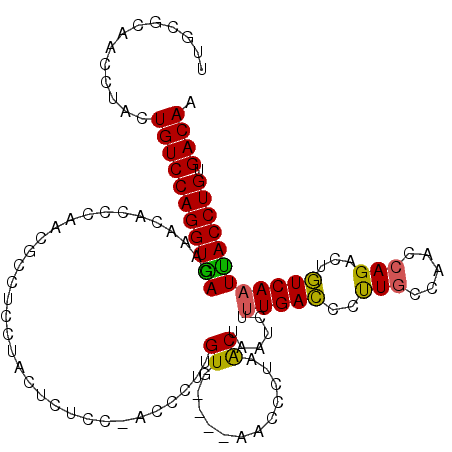
| Structure conservation index | 0.79 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15922224 120 + 23771897 AGUGGCUAACCCGCCAGCCCACUUGUCCGGUGGAUCGAAAUUCCCUGACUACUGCCAACUUGCGGGCAGUGCCUCGCAUAUUGCGCAACCUACUGUCCAGGUGAGAAAAACCCAACGCCU ..((((......))))...((((((..((((((.(((........)))....((((((..(((((((...))).))))..))).)))..))))))..))))))................. ( -36.80) >DroVir_CAF1 3605 106 + 1 AAUGGCUAACACGCCAGCCCACAUGUCCCGUAGAUCGCAAUGCACUAACUACCGCGAAUUUGCGAGCAGUGCCGCGCAUAUUGCGCAACUUGUUGUCCAGGUAGGU-------------- ..((((......))))((((((.(((..((((((((((...............))).))))))).))))))..((((.....)))).(((((.....))))).)))-------------- ( -30.56) >DroSim_CAF1 24267 120 + 1 AGUGGCUAACCCGCCAGCCCACUUGUCCGGUGGAUCGCAAUGCCUUGACCACUGCCAACUUGCGAGCAGUGCCUCGCAUAUUGCGCAACCUACUGUCCAGGUGAGAAACACCCAACGCCU ..((((......))))...((((((..((((((..(((((((...(((.(((((((.......).))))))..)))..)))))))....))))))..))))))................. ( -37.30) >DroEre_CAF1 21606 120 + 1 AGUGGCUAACCCGCCAGCCCACUUGUCCGGUGGAUCGCAAUGCCCUGACAACUGCCAACUUGCGGGCAGUUCCUCGCAUAUUGCGCAACCUACUGUCCAGGUGAGAACCACCCAACGCCU ..((((......))))...((((((..((((((..(((((((...(((.(((((((........)))))))..)))..)))))))....))))))..))))))................. ( -39.40) >DroYak_CAF1 22212 120 + 1 AGUGGCUAACCCGCCAGCCCACCUGUCCGGUGGAUCGCAAUGCCCUGACUACUGCCAACUUGCGGGCAGUGCCUCGCAUAUUGCGCAACCUACUGUCCAGGUGAGAACCAACCAACCUAC ..((((......))))...((((((..((((((..(((((((...(((.(((((((........)))))))..)))..)))))))....))))))..))))))................. ( -42.20) >DroMoj_CAF1 2004 106 + 1 AAUGGCUGACACGCCAGCCCACAUGCCCAGUUGAUCGUAAUGCCCUAACUACUGCAAAUCUGCGAGCUGUGCCGCGCAUAUUGCGCAACUUGUUGUCCAGGUAACU-------------- ...(((((......))))).....((.((((...(((((.(((..........)))....))))))))).)).((((.....)))).(((((.....)))))....-------------- ( -27.70) >consensus AGUGGCUAACCCGCCAGCCCACUUGUCCGGUGGAUCGCAAUGCCCUGACUACUGCCAACUUGCGAGCAGUGCCUCGCAUAUUGCGCAACCUACUGUCCAGGUGAGAA_CACCCAACGCCU ..((((......))))...((((((..((((((..(((((((.......((((((..........)))))).......)))))))....))))))..))))))................. (-28.21 = -28.41 + 0.20)



| Location | 15,922,304 – 15,922,418 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -20.03 |
| Consensus MFE | -16.03 |
| Energy contribution | -15.45 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15922304 114 + 23771897 UUGCGCAACCUACUGUCCAGGUGAGAAAAACCCAACGCCUUCUACUCUGUGAACCUUGUG----GACUCUAACUAAUCUUUGACCCUUGCCAACCAGACUGUCAAUCACCUGUGACAA .............(((((((((((................(((((...(....)...)))----)).............(((((..(((.....)))...)))))))))))).)))). ( -23.90) >DroSec_CAF1 24016 113 + 1 UUGCGCAACCUACUGUCCAGGUGAGAAACACCCAACGCCUCCUACUCUAC-GCCCUUGUA----AACCCUAACUAAUCUUUGACCCUUGCCAACCAGACUGUCAAUUACCUGUGACAA .............(((((((((((.......................(((-(....))))----...............(((((..(((.....)))...)))))))))))).)))). ( -17.20) >DroSim_CAF1 24347 113 + 1 UUGCGCAACCUACUGUCCAGGUGAGAAACACCCAACGCCUCCUACUCUCC-ACCCUUGUG----AACCGUAACUAAUCUUUGACCCUUGCCAACCAGACUGUCAAUUACCUGUGACAA .............(((((((((((..................(((....(-((....)))----....)))........(((((..(((.....)))...)))))))))))).)))). ( -18.40) >DroEre_CAF1 21686 113 + 1 UUGCGCAACCUACUGUCCAGGUGAGAACCACCCAACGCCUUUAACUCUCC-ACCCUUGUG----AAACCUAACUAAUCUUUGACCCCUGCCAACCAGACUGUCAAUUACCUGUGACAA .............(((((((((((.........................(-((....)))----...............(((((..(((.....)))...)))))))))))).)))). ( -20.40) >DroYak_CAF1 22292 113 + 1 UUGCGCAACCUACUGUCCAGGUGAGAACCAACCAACCUACUUAACUCUCC-ACCCUUGUG----AGCCCUAACUAAUCUUUGACCCCUGCCCACCAGACUGUCAAUUACCUGUGACAA .............(((((((((((......................((((-(....)).)----)).............(((((..(((.....)))...)))))))))))).)))). ( -20.70) >DroMoj_CAF1 2084 97 + 1 UUGCGCAACUUGUUGUCCAGGUAACU--------------CUUUUGCACG-ACUCUUGUUUUUCAUACCUAGCUAA----UGAGCGUUUUCA--CAGGCUCUCAAUUACCUGCGACAA ............((((((((((((..--------------.........(-((....)))................----((((.(((....--..))).)))).))))))).))))) ( -19.60) >consensus UUGCGCAACCUACUGUCCAGGUGAGAAACACCCAACGCCUCCUACUCUCC_ACCCUUGUG____AACCCUAACUAAUCUUUGACCCUUGCCAACCAGACUGUCAAUUACCUGUGACAA .............(((((((((((.................................((............))......(((((..(((.....)))...)))))))))))).)))). (-16.03 = -15.45 + -0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:48 2006