
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,920,485 – 15,920,645 |
| Length | 160 |
| Max. P | 0.990340 |

| Location | 15,920,485 – 15,920,605 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -36.69 |
| Consensus MFE | -34.48 |
| Energy contribution | -34.04 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
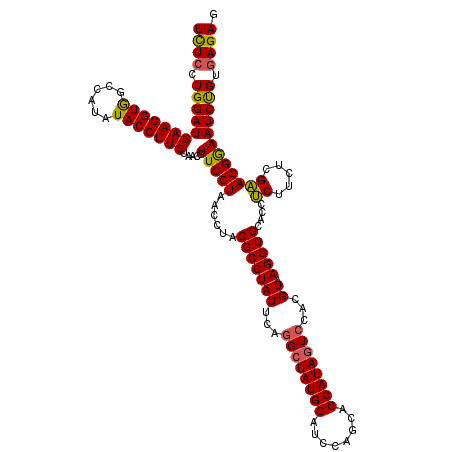
>3L_DroMel_CAF1 15920485 120 + 23771897 UCUCUUGGAUGAAGGUGGCCAUAUACCUUCUUACGUUCCUAACCUACGCCUAUUCAGGCUAUGGAUCCAGCACCAUAGUCCACGUAAGGUGCACCGCUUCUCGUAGGAAAUCUGUGAGAG ((((.((((((((((((......))))))).....((((((.((...((((....))))...)).....(((((.(((....).)).)))))...........))))))))))).)))). ( -37.10) >DroSec_CAF1 22173 120 + 1 UCUCCUGGAUGAAGGUGGCCAUAUACCUUCUAACGUUCCUAACCUACGCCUAUUCAGGCUAUGGAUCCAGCACCAUAGUCCACGUAAGGUGCACCUCUUCUCGAAGGGAAUCUGUGAGAG ((((.((((((((((((......))))))).....(((((..((...((((....))))...)).....(((((.(((....).)).)))))............)))))))))).)))). ( -35.30) >DroSim_CAF1 22502 120 + 1 UCUCCUGGAUGAAGGUGGCCAUAUACCUUCUAACGUUCCUAACCUACGCCUAUUCAGGCUAUGGAUCCAGCACCAUAGUCCACGUAAGGUGCACCUCUUCUCGAAGGGAAUCUGUGAGAG ((((.((((((((((((......))))))).....(((((..((...((((....))))...)).....(((((.(((....).)).)))))............)))))))))).)))). ( -35.30) >DroEre_CAF1 19843 120 + 1 UCUCGUGGAUGAAGGUAGCCAUAUACCUUCUAACGUUCCUAACGUACGCCUAUUCAGGGUAUGGAUCCAGCACCAUAGUCCAUGUAAGGUGCACCUCUUUUCGAAGGGAAUCCGUGAGAG (((((((((((((((((......))))))).....(((((..(((((.(((....))))))))......(((((.............)))))............))))))))))))))). ( -40.42) >DroYak_CAF1 20483 120 + 1 UUUCGUGGAUGAAGGUGGCCAUAUACCUUCUAACGUUCCUGACAUACGCCUAUUCAGGCUAUGGAUCCAGCACCAUAGUACAUGUAAGGUGCACCUCUUUUCGAAGGGAAUCCGUAAGAG (((..((((((((((((......))))))).....(((((..((((.((((....))))))))......(((((.............)))))............))))))))))..))). ( -35.32) >consensus UCUCCUGGAUGAAGGUGGCCAUAUACCUUCUAACGUUCCUAACCUACGCCUAUUCAGGCUAUGGAUCCAGCACCAUAGUCCACGUAAGGUGCACCUCUUCUCGAAGGGAAUCUGUGAGAG ((((.((((((((((((......))))))).....(((((......(((((((...((((((((........))))))))...)).)))))....((.....)))))))))))).)))). (-34.48 = -34.04 + -0.44)

| Location | 15,920,525 – 15,920,645 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -27.48 |
| Energy contribution | -28.16 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
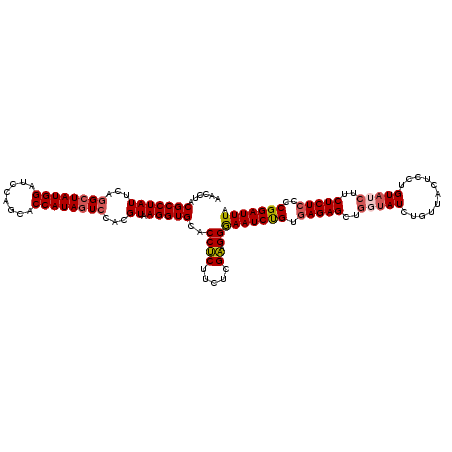
>3L_DroMel_CAF1 15920525 120 + 23771897 AACCUACGCCUAUUCAGGCUAUGGAUCCAGCACCAUAGUCCACGUAAGGUGCACCGCUUCUCGUAGGAAAUCUGUGAGAGCUAGUAUCUGUUACUCCUGUAUCUUCUCUCCCCGGAUUUA ..(((((((((((...((((((((........))))))))...)).))))((...)).....)))))(((((((.(((((...((((..(.....)..))))...)))))..))))))). ( -32.50) >DroSec_CAF1 22213 120 + 1 AACCUACGCCUAUUCAGGCUAUGGAUCCAGCACCAUAGUCCACGUAAGGUGCACCUCUUCUCGAAGGGAAUCUGUGAGAGCUGGUAUCUGUUACUCCUGUAUCUUCUCUCCCCGGAUUUA ......(((((((...((((((((........))))))))...)).)))))..(((........)))(((((((.(((((..(((((..(.....)..)))))..)))))..))))))). ( -33.60) >DroSim_CAF1 22542 120 + 1 AACCUACGCCUAUUCAGGCUAUGGAUCCAGCACCAUAGUCCACGUAAGGUGCACCUCUUCUCGAAGGGAAUCUGUGAGAGCUGGUAUCUGUUACUCCUGUAUCUUCUCUCCCCGGAUUUA ......(((((((...((((((((........))))))))...)).)))))..(((........)))(((((((.(((((..(((((..(.....)..)))))..)))))..))))))). ( -33.60) >DroEre_CAF1 19883 119 + 1 AACGUACGCCUAUUCAGGGUAUGGAUCCAGCACCAUAGUCCAUGUAAGGUGCACCUCUUUUCGAAGGGAAUCCGUGAGAGAUGCUAUCUG-UACUCCUGUAUCUUCUCUCCCCGGAUUUA ..(((((.(((....))))))))......(((((.............))))).(((........)))(((((((.(((((((((......-.......))))).))))....))))))). ( -33.94) >DroYak_CAF1 20523 119 + 1 GACAUACGCCUAUUCAGGCUAUGGAUCCAGCACCAUAGUACAUGUAAGGUGCACCUCUUUUCGAAGGGAAUCCGUAAGAGAUGCCAUAUG-UACUCCUGUAUCUUCUCUCCCCGGAUUUA ..((((.((((....))))))))(((((.(((((.............)))))..........((((((.((((....).))).))....(-(((....)))))))).......))))).. ( -31.52) >consensus AACCUACGCCUAUUCAGGCUAUGGAUCCAGCACCAUAGUCCACGUAAGGUGCACCUCUUCUCGAAGGGAAUCUGUGAGAGCUGGUAUCUGUUACUCCUGUAUCUUCUCUCCCCGGAUUUA ......(((((((...((((((((........))))))))...)).)))))..((((.....)).))(((((((.(((((..(((((...........)))))..)))))..))))))). (-27.48 = -28.16 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:46 2006