
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,919,427 – 15,919,581 |
| Length | 154 |
| Max. P | 0.999978 |

| Location | 15,919,427 – 15,919,547 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -38.99 |
| Consensus MFE | -33.10 |
| Energy contribution | -33.34 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15919427 120 + 23771897 CUGUUUCGGUUUUAAGCAAACAGCGAGCGCGCCUCUUUAACGGUUUUCGGGUGUUGCGUUCGCAUGUUUAUGCACGUUGAUUUCGCUGGCCGUUGUAAUAAAGGCCAACAAGGUCAGGUG ...............((((((((((((((((((((.............))).).))))))))).))))..)))((.(((((((.(.(((((...........))))).).))))))))). ( -36.12) >DroSec_CAF1 21132 120 + 1 CUGUUUCGGUUUUAAGCAAACAACGAGUGCGCCUCUUUAACGGUUUUCGGGUGUUGCGUUCGCAUGUUUAUGCACGUUGAUUUCGCUGGCCGUUGUAAUAAAGGCCAACAAGGUCAGGUG .((((((........).))))).......(((((...((((((((..((((....((((..((((....))))))))....))))..)))))))).......((((.....))))))))) ( -30.70) >DroSim_CAF1 21451 120 + 1 CUGUUUCGGUUUUAAGCAAACAGCGAGCGCGCCUCUUUAACGGUUUUCGGGUGUUGCGUUCGCAUGUUUAUGCACGUUGAUUUCGCUGGCCGUUGUAAUAAAGGCCAACAAGGUCAGGUG ...............((((((((((((((((((((.............))).).))))))))).))))..)))((.(((((((.(.(((((...........))))).).))))))))). ( -36.12) >DroEre_CAF1 18797 117 + 1 CUGUUUCGGUU-UAAGCAAACAGCGAGCGCG--UCUUUAACGGUUUUCGGGUGUUGCGUUCGCAUGUUUGUGCACGUUGAUUUUGUUGGCCGCUGUAAUAAAGGCCAACAAGGUCAGGUG ...........-...((((((((((((((((--.(.....((.....))...).))))))))).))))))).(((.(((((((((((((((...........)))))))))))))))))) ( -47.60) >DroYak_CAF1 19370 119 + 1 CUGUUUCGGUU-UAAGCAAACAGCGAGCGCGCCUCUUUAACGGUUUUCGGGUGUUGCGUUCGCAUGUUUAUGCACGUUGAUUUUGUUGGCCGUUGUAAUAAAGGCCAACAAGGUCAGGUG ...........-...((((((((((((((((((((.............))).).))))))))).))))..)))((.(((((((((((((((...........))))))))))))))))). ( -44.42) >consensus CUGUUUCGGUUUUAAGCAAACAGCGAGCGCGCCUCUUUAACGGUUUUCGGGUGUUGCGUUCGCAUGUUUAUGCACGUUGAUUUCGCUGGCCGUUGUAAUAAAGGCCAACAAGGUCAGGUG ...............((((((((((((((((.(.((..((.....))..)).).))))))))).))))..)))((.(((((((.(.(((((...........))))).).))))))))). (-33.10 = -33.34 + 0.24)



| Location | 15,919,427 – 15,919,547 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -29.18 |
| Energy contribution | -28.98 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15919427 120 - 23771897 CACCUGACCUUGUUGGCCUUUAUUACAACGGCCAGCGAAAUCAACGUGCAUAAACAUGCGAACGCAACACCCGAAAACCGUUAAAGAGGCGCGCUCGCUGUUUGCUUAAAACCGAAACAG (((.(((..(((((((((...........)))))))))..)))..)))..((((((.((((.(((..(....)....((........)).))).))))))))))................ ( -30.80) >DroSec_CAF1 21132 120 - 1 CACCUGACCUUGUUGGCCUUUAUUACAACGGCCAGCGAAAUCAACGUGCAUAAACAUGCGAACGCAACACCCGAAAACCGUUAAAGAGGCGCACUCGUUGUUUGCUUAAAACCGAAACAG (((.(((..(((((((((...........)))))))))..)))..))).........((((..(((((...((.....)).....(((.....))))))))))))............... ( -27.50) >DroSim_CAF1 21451 120 - 1 CACCUGACCUUGUUGGCCUUUAUUACAACGGCCAGCGAAAUCAACGUGCAUAAACAUGCGAACGCAACACCCGAAAACCGUUAAAGAGGCGCGCUCGCUGUUUGCUUAAAACCGAAACAG (((.(((..(((((((((...........)))))))))..)))..)))..((((((.((((.(((..(....)....((........)).))).))))))))))................ ( -30.80) >DroEre_CAF1 18797 117 - 1 CACCUGACCUUGUUGGCCUUUAUUACAGCGGCCAACAAAAUCAACGUGCACAAACAUGCGAACGCAACACCCGAAAACCGUUAAAGA--CGCGCUCGCUGUUUGCUUA-AACCGAAACAG (((.(((..(((((((((...........)))))))))..)))..)))..((((((.((((.(((.......(.....)(((...))--)))).))))))))))....-........... ( -31.70) >DroYak_CAF1 19370 119 - 1 CACCUGACCUUGUUGGCCUUUAUUACAACGGCCAACAAAAUCAACGUGCAUAAACAUGCGAACGCAACACCCGAAAACCGUUAAAGAGGCGCGCUCGCUGUUUGCUUA-AACCGAAACAG (((.(((..(((((((((...........)))))))))..)))..)))..((((((.((((.(((..(....)....((........)).))).))))))))))....-........... ( -31.10) >consensus CACCUGACCUUGUUGGCCUUUAUUACAACGGCCAGCGAAAUCAACGUGCAUAAACAUGCGAACGCAACACCCGAAAACCGUUAAAGAGGCGCGCUCGCUGUUUGCUUAAAACCGAAACAG (((.(((..(((((((((...........)))))))))..)))..)))..((((((.((((.(((.......(.....)(((.....)))))).))))))))))................ (-29.18 = -28.98 + -0.20)



| Location | 15,919,467 – 15,919,581 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.88 |
| Mean single sequence MFE | -47.42 |
| Consensus MFE | -42.92 |
| Energy contribution | -43.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15919467 114 + 23771897 CGGUUUUCGGGUGUUGCGUUCGCAUGUUUAUGCACGUUGAUUUCGCUGGCCGUUGUAAUAAAGGCCAACAAGGUCAGGUGCAAACGAAAUGCGAGU------ACGAGUGACGCCUGCGUG .......(((((((..(.(((((((.....(((((.(((((((.(.(((((...........))))).).))))))))))))......))))))).------....)..))))))).... ( -43.50) >DroSec_CAF1 21172 114 + 1 CGGUUUUCGGGUGUUGCGUUCGCAUGUUUAUGCACGUUGAUUUCGCUGGCCGUUGUAAUAAAGGCCAACAAGGUCAGGUGCAAACGAAAUGCGAGU------ACGAGUGACGCCUGCGUG .......(((((((..(.(((((((.....(((((.(((((((.(.(((((...........))))).).))))))))))))......))))))).------....)..))))))).... ( -43.50) >DroSim_CAF1 21491 114 + 1 CGGUUUUCGGGUGUUGCGUUCGCAUGUUUAUGCACGUUGAUUUCGCUGGCCGUUGUAAUAAAGGCCAACAAGGUCAGGUGCAAACGAAAUGCGAGU------ACGAGUGACGCCUGCGUG .......(((((((..(.(((((((.....(((((.(((((((.(.(((((...........))))).).))))))))))))......))))))).------....)..))))))).... ( -43.50) >DroEre_CAF1 18834 114 + 1 CGGUUUUCGGGUGUUGCGUUCGCAUGUUUGUGCACGUUGAUUUUGUUGGCCGCUGUAAUAAAGGCCAACAAGGUCAGGUGCAAACGAAUUGCGAGU------ACGAGUGACGCCUGCGUG .......(((((((..(.((((((.((((((((((.(((((((((((((((...........)))))))))))))))))))..)))))))))))).------....)..))))))).... ( -52.70) >DroYak_CAF1 19409 120 + 1 CGGUUUUCGGGUGUUGCGUUCGCAUGUUUAUGCACGUUGAUUUUGUUGGCCGUUGUAAUAAAGGCCAACAAGGUCAGGUGCAAACGAAAUGCGAGUACGAGUACGAGUGACGCCUGCGUG .......(((((((..(..((((((.....(((((.(((((((((((((((...........))))))))))))))))))))......))))))(((....)))..)..))))))).... ( -53.90) >consensus CGGUUUUCGGGUGUUGCGUUCGCAUGUUUAUGCACGUUGAUUUCGCUGGCCGUUGUAAUAAAGGCCAACAAGGUCAGGUGCAAACGAAAUGCGAGU______ACGAGUGACGCCUGCGUG .......(((((((..(..((((((.....(((((.(((((((.(.(((((...........))))).).))))))))))))......))))))(((....)))..)..))))))).... (-42.92 = -43.32 + 0.40)


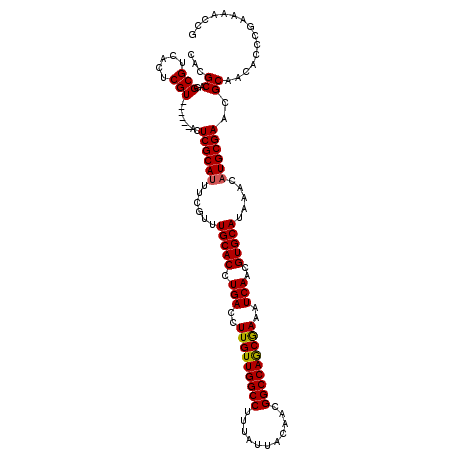
| Location | 15,919,467 – 15,919,581 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.88 |
| Mean single sequence MFE | -33.94 |
| Consensus MFE | -33.24 |
| Energy contribution | -32.96 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.18 |
| SVM RNA-class probability | 0.999978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15919467 114 - 23771897 CACGCAGGCGUCACUCGU------ACUCGCAUUUCGUUUGCACCUGACCUUGUUGGCCUUUAUUACAACGGCCAGCGAAAUCAACGUGCAUAAACAUGCGAACGCAACACCCGAAAACCG ......((.((....(((------..((((((......(((((.(((..(((((((((...........)))))))))..)))..))))).....)))))))))....))))........ ( -33.90) >DroSec_CAF1 21172 114 - 1 CACGCAGGCGUCACUCGU------ACUCGCAUUUCGUUUGCACCUGACCUUGUUGGCCUUUAUUACAACGGCCAGCGAAAUCAACGUGCAUAAACAUGCGAACGCAACACCCGAAAACCG ......((.((....(((------..((((((......(((((.(((..(((((((((...........)))))))))..)))..))))).....)))))))))....))))........ ( -33.90) >DroSim_CAF1 21491 114 - 1 CACGCAGGCGUCACUCGU------ACUCGCAUUUCGUUUGCACCUGACCUUGUUGGCCUUUAUUACAACGGCCAGCGAAAUCAACGUGCAUAAACAUGCGAACGCAACACCCGAAAACCG ......((.((....(((------..((((((......(((((.(((..(((((((((...........)))))))))..)))..))))).....)))))))))....))))........ ( -33.90) >DroEre_CAF1 18834 114 - 1 CACGCAGGCGUCACUCGU------ACUCGCAAUUCGUUUGCACCUGACCUUGUUGGCCUUUAUUACAGCGGCCAACAAAAUCAACGUGCACAAACAUGCGAACGCAACACCCGAAAACCG ......((.((....(((------..(((((.......(((((.(((..(((((((((...........)))))))))..)))..)))))......))))))))....))))........ ( -32.22) >DroYak_CAF1 19409 120 - 1 CACGCAGGCGUCACUCGUACUCGUACUCGCAUUUCGUUUGCACCUGACCUUGUUGGCCUUUAUUACAACGGCCAACAAAAUCAACGUGCAUAAACAUGCGAACGCAACACCCGAAAACCG .......((((.....(((....)))((((((......(((((.(((..(((((((((...........)))))))))..)))..))))).....))))))))))............... ( -35.80) >consensus CACGCAGGCGUCACUCGU______ACUCGCAUUUCGUUUGCACCUGACCUUGUUGGCCUUUAUUACAACGGCCAGCGAAAUCAACGUGCAUAAACAUGCGAACGCAACACCCGAAAACCG ...((..(((.....)))........((((((......(((((.(((..(((((((((...........)))))))))..)))..))))).....))))))..))............... (-33.24 = -32.96 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:43 2006