
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,913,568 – 15,913,871 |
| Length | 303 |
| Max. P | 0.988452 |

| Location | 15,913,568 – 15,913,684 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.39 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -17.70 |
| Energy contribution | -17.54 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15913568 116 + 23771897 GGCCCGGUCCAAUCAUUGUAGUGUUCGUAAAUUGAAAGUGAAAUGGAAUGGCCGUCGGUCAGAUGUGUGUGUGGUACUAUAUGUUCAUACAUGU----GUACUACUAUGCUUCGAGCAAC ((((...((((.(((((.((((........))))..)))))..))))..))))...(.((.((.(((((.(((((((.((((((....))))))----)))))))))))).)))).)... ( -36.30) >DroSec_CAF1 15314 98 + 1 GGCCCGGUCCACUCAUUGUAGUGUUCGUAAAUUGAAAGUGAAAUGGAAUGGCCGCCGGUCAGAU------------------GUUCAUACAUAU----GUACUACUAUGCAUCGAGCAAC ((((...((((.(((((.((((........))))..)))))..))))..))))((((((((...------------------((...(((....----)))..))..)).)))).))... ( -24.50) >DroSim_CAF1 15515 102 + 1 GGCCCGGUCCACUCAUCGUAGUGUUCGUAAAUUGAAAGUGAAAUGGAAUGGCCGCCGGUCAGAU------------------GUUCAUACAUAUGUAUGUACUACUAUGCAUCGAGCAAC ((((...((((.((((..((((........))))...))))..))))..))))((((((((...------------------((.(((((....))))).)).....)).)))).))... ( -28.60) >DroEre_CAF1 12982 110 + 1 GGCCCGGUCUAGUCAUUGUAGUGCUCGUAAAUUGAAAGUGAAAUGGAAUGGCCGCUGGUACUACAAGUA--UACAAGU----AUAUGUACAUAU----GUACUGCUGUGCAGCCAGCAAC ((((...((((.(((((.((((........))))..)))))..))))..))))((((((..((((.(((--(....))----)).))))....(----((((....)))))))))))... ( -33.30) >DroYak_CAF1 13145 107 + 1 GGCCCGGUCCACUCAUUGUAGUGUUCGUAAAUUGAAAGUGAAAUGGAAUGGCCGCUGGUACUACACUCG--UAGGUAC----AUAUAUACAUAU----GUAC---AGUGCAUCCAGCAAC ((((...((((.(((((.((((........))))..)))))..))))..))))(((((..((((....)--)))((((----((((....))))----))))---.......)))))... ( -35.90) >consensus GGCCCGGUCCACUCAUUGUAGUGUUCGUAAAUUGAAAGUGAAAUGGAAUGGCCGCCGGUCAGAU_______U__________GUUCAUACAUAU____GUACUACUAUGCAUCGAGCAAC ((((...((((.((((..((((........))))...))))..))))..))))......................................................(((.....))).. (-17.70 = -17.54 + -0.16)



| Location | 15,913,608 – 15,913,724 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -14.68 |
| Energy contribution | -14.68 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15913608 116 + 23771897 AAAUGGAAUGGCCGUCGGUCAGAUGUGUGUGUGGUACUAUAUGUUCAUACAUGU----GUACUACUAUGCUUCGAGCAACAAGUGUUUUAUCAACAGAGCGACAGGCGUCCGAACAAAAG ...((((...(((((((.((.((.(((((.(((((((.((((((....))))))----)))))))))))).))(((((.....)))))........)).)))).))).))))........ ( -37.90) >DroSec_CAF1 15354 98 + 1 AAAUGGAAUGGCCGCCGGUCAGAU------------------GUUCAUACAUAU----GUACUACUAUGCAUCGAGCAACAAGUGUUUUAUCAACUGAGCGACAGGCGUCCGGACAAAAG ...((((...(((..((.((((.(------------------(.....((((.(----((.......(((.....)))))).)))).....)).)))).))...))).))))........ ( -24.21) >DroSim_CAF1 15555 102 + 1 AAAUGGAAUGGCCGCCGGUCAGAU------------------GUUCAUACAUAUGUAUGUACUACUAUGCAUCGAGCAACAAGUGUUUUAUCAACUGAGCGACAGGCGUCCGGACAAAAG ...((...(((.((((.(((.(((------------------((.(((((....))))).........)))))..((....(((.........)))..))))).)))).)))..)).... ( -27.00) >DroEre_CAF1 13022 110 + 1 AAAUGGAAUGGCCGCUGGUACUACAAGUA--UACAAGU----AUAUGUACAUAU----GUACUGCUGUGCAGCCAGCAACAAGUGUUUUAUCAACACAGAGACAGGCGUCCGGACAAAAG ...((((...(((((((((..((((.(((--(....))----)).))))....(----((((....))))))))))).....(((((.....))))).......))).))))........ ( -32.30) >DroYak_CAF1 13185 107 + 1 AAAUGGAAUGGCCGCUGGUACUACACUCG--UAGGUAC----AUAUAUACAUAU----GUAC---AGUGCAUCCAGCAACAAGUGUUUUAUCAACACAGAGACAGGCGUCCGGACAAAAG ...((((...((((((((..((((....)--)))((((----((((....))))----))))---.......))))).....(((((.....))))).......))).))))........ ( -33.20) >consensus AAAUGGAAUGGCCGCCGGUCAGAU_______U__________GUUCAUACAUAU____GUACUACUAUGCAUCGAGCAACAAGUGUUUUAUCAACAGAGCGACAGGCGUCCGGACAAAAG ...((...(((.((((.(((...........................(((........)))......(((.....)))......................))).)))).)))..)).... (-14.68 = -14.68 + -0.00)


| Location | 15,913,608 – 15,913,724 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -14.74 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15913608 116 - 23771897 CUUUUGUUCGGACGCCUGUCGCUCUGUUGAUAAAACACUUGUUGCUCGAAGCAUAGUAGUAC----ACAUGUAUGAACAUAUAGUACCACACACACAUCUGACCGACGGCCAUUCCAUUU .........(((.(((.((((.((...((.............(((.....)))..((.((((----..((((....))))...)))).)).....))...)).)))))))...))).... ( -23.60) >DroSec_CAF1 15354 98 - 1 CUUUUGUCCGGACGCCUGUCGCUCAGUUGAUAAAACACUUGUUGCUCGAUGCAUAGUAGUAC----AUAUGUAUGAAC------------------AUCUGACCGGCGGCCAUUCCAUUU .........(((.(((.((((.((((.((.......(((...(((.....))).))).((((----....))))...)------------------).)))).)))))))...))).... ( -25.20) >DroSim_CAF1 15555 102 - 1 CUUUUGUCCGGACGCCUGUCGCUCAGUUGAUAAAACACUUGUUGCUCGAUGCAUAGUAGUACAUACAUAUGUAUGAAC------------------AUCUGACCGGCGGCCAUUCCAUUU .........(((.(((.((((.((((.((.....(((((...(((.....))).))).)).(((((....)))))..)------------------).)))).)))))))...))).... ( -28.60) >DroEre_CAF1 13022 110 - 1 CUUUUGUCCGGACGCCUGUCUCUGUGUUGAUAAAACACUUGUUGCUGGCUGCACAGCAGUAC----AUAUGUACAUAU----ACUUGUA--UACUUGUAGUACCAGCGGCCAUUCCAUUU .........(((.(((.......(((((.....))))).....(((((((((...))))(((----(..((((((...----...))))--))..))))...))))))))...))).... ( -32.30) >DroYak_CAF1 13185 107 - 1 CUUUUGUCCGGACGCCUGUCUCUGUGUUGAUAAAACACUUGUUGCUGGAUGCACU---GUAC----AUAUGUAUAUAU----GUACCUA--CGAGUGUAGUACCAGCGGCCAUUCCAUUU .........(((.(((.......(((((.....))))).....(((((.((((((---((((----((((....))))----))))...--..))))))...))))))))...))).... ( -33.90) >consensus CUUUUGUCCGGACGCCUGUCGCUCUGUUGAUAAAACACUUGUUGCUCGAUGCAUAGUAGUAC____AUAUGUAUGAAC__________A_______AUCUGACCGGCGGCCAUUCCAUUU .........(((.(((.((((.((.(((.....)))(((...(((.....)))....)))........................................)).)))))))...))).... (-14.74 = -15.10 + 0.36)

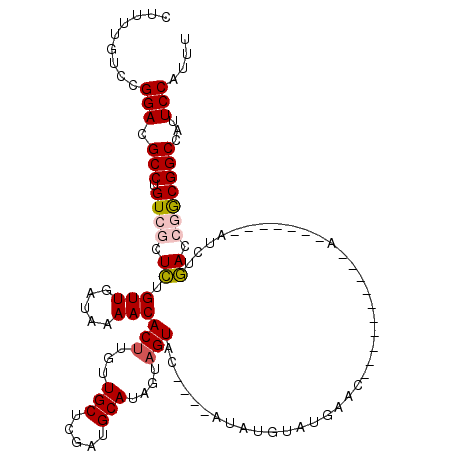

| Location | 15,913,724 – 15,913,839 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.04 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -28.70 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15913724 115 - 23771897 UUGUUAUUAAUAAUAUUAGUA-----UGUGAUAUUAACGGGACUGCAGCCUCCAUUGUGCCGGCGGUGGAAACCCUUUGGCACGCGAACUCAAUUUUCCUAGGCUGACAGUUUAUAAUUC ..........((((((((...-----..))))))))...((((((((((((.....(((((((.((.(....))).)))))))..(((.......)))..)))))).))))))....... ( -34.60) >DroSec_CAF1 15452 115 - 1 UUGUUAUUAAUAAUAUUAGUA-----UGUGAUAUUAACGGGACUGCAGCCUCCAUUGUGUCGGCGGUGGAAACCCUUUGGCACGCGAACUCAAUUUUCCUAGGCUGACAGUUUAGAAUGC ..........((((((((...-----..)))))))).(..(((((((((((.....(((((((.((.(....))).)))))))..(((.......)))..)))))).)))))..)..... ( -32.40) >DroSim_CAF1 15657 115 - 1 UUGUUAUUAAUAAUAUUAGUA-----UGUGAUAUUAACGGGACUGCAGCCUCCAUUGUGCCGGCGGUGGAAACCCUUUGGCACGCGAACUCAAUUUUCCUAGGCUGACAGUUUAUAAUGC ....(((((.((((((((...-----..))))))))...((((((((((((.....(((((((.((.(....))).)))))))..(((.......)))..)))))).)))))).))))). ( -35.10) >DroEre_CAF1 13132 115 - 1 UUGUUAUUAAUAAUAUUAGUA-----UGUGAUAUUAACUGGACUGCAGCCUCCUUUGUGGCGGCGGUGGAAACCCUUUGGCAUGCGAACUCAAUUUUCCUAGGCUGACAGUUUAUAAUGC ....(((((.((((((((...-----..))))))))..(((((((((((((.....(((.(((.((.(....))).))).)))..(((.......)))..)))))).)))))))))))). ( -29.00) >DroYak_CAF1 13292 120 - 1 UUGUUAUUAAUAAUAUUAGUACGAGAUGUGAUAUUAACUGGACUGCAGCCUCCUUUCUGGCGGCGGUGGAAACCCUUUGGCAUGCGAACUCAAUUUUCCUAGGCUGACAGUUUAUAAUGC ....(((((.((((((((..........))))))))..(((((((((((((......((.(((.((.(....))).))).))...(((.......)))..)))))).)))))))))))). ( -26.50) >consensus UUGUUAUUAAUAAUAUUAGUA_____UGUGAUAUUAACGGGACUGCAGCCUCCAUUGUGCCGGCGGUGGAAACCCUUUGGCACGCGAACUCAAUUUUCCUAGGCUGACAGUUUAUAAUGC ..........((((((((..........))))))))...((((((((((((.....(((((((.((.(....))).)))))))..(((.......)))..)))))).))))))....... (-28.70 = -29.10 + 0.40)



| Location | 15,913,764 – 15,913,871 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.00 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -25.83 |
| Energy contribution | -26.19 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15913764 107 + 23771897 CCAAAGGGUUUCCACCGCCGGCACAAUGGAGGCUGCAGUCCCGUUAAUAUCACA-----UACUAAUAUUAUUAAUAACAAUGGCUGCAGUCUUGAUUCAAUUUGAUGGCUG--------C ......(((....)))((((.((.(((.((((((((((((..(((((((.....-----.........)))))))......)))))))))))).))).....)).))))..--------. ( -28.64) >DroSec_CAF1 15492 107 + 1 CCAAAGGGUUUCCACCGCCGACACAAUGGAGGCUGCAGUCCCGUUAAUAUCACA-----UACUAAUAUUAUUAAUAACAAUGGCUGCAGUCUUGAUUCAAUUUGAUGGCUG--------C ......(((....)))((((.((.(((.((((((((((((..(((((((.....-----.........)))))))......)))))))))))).))).....)).))))..--------. ( -28.64) >DroSim_CAF1 15697 107 + 1 CCAAAGGGUUUCCACCGCCGGCACAAUGGAGGCUGCAGUCCCGUUAAUAUCACA-----UACUAAUAUUAUUAAUAACAAUGGCUGCAGUCUUGAUUCAAUUUGAUGGCUG--------C ......(((....)))((((.((.(((.((((((((((((..(((((((.....-----.........)))))))......)))))))))))).))).....)).))))..--------. ( -28.64) >DroEre_CAF1 13172 103 + 1 CCAAAGGGUUUCCACCGCCGCCACAAAGGAGGCUGCAGUCCAGUUAAUAUCACA-----UACUAAUAUUAUUAAUAACAAUGACUGCAGUCUUGAUUCAAUUUGAUGG------------ .((((..(..((.....((........))(((((((((((..(((((((.....-----.........)))))))......))))))))))).))..)..))))....------------ ( -23.94) >DroYak_CAF1 13332 120 + 1 CCAAAGGGUUUCCACCGCCGCCAGAAAGGAGGCUGCAGUCCAGUUAAUAUCACAUCUCGUACUAAUAUUAUUAAUAACAAUGACUGCAGUCUUGAUUCAAUUUGAUGGCUACCCGGUUGC .(((.((((..(((.((......(((..((((((((((((..(((((((...................)))))))......))))))))))))..)))....)).)))..))))..))). ( -32.51) >consensus CCAAAGGGUUUCCACCGCCGCCACAAUGGAGGCUGCAGUCCCGUUAAUAUCACA_____UACUAAUAUUAUUAAUAACAAUGGCUGCAGUCUUGAUUCAAUUUGAUGGCUG________C ......(((....)))((((.((.(((.((((((((((((..(((((((...................)))))))......)))))))))))).))).....)).))))........... (-25.83 = -26.19 + 0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:29 2006