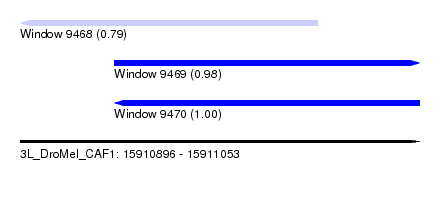
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,910,896 – 15,911,053 |
| Length | 157 |
| Max. P | 0.998939 |

| Location | 15,910,896 – 15,911,013 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
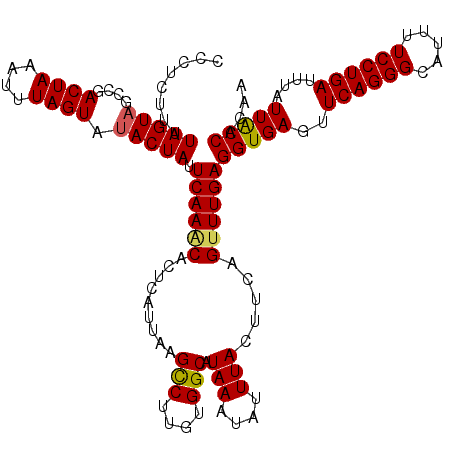
>3L_DroMel_CAF1 15910896 117 - 23771897 ACACUCAUUAAGUCUUGUGGCAUAAAUAUUUACUUCAGUUUGAGGUGAGUUCAGGGCAUUUUCCUGAUAUAUUACCACAACAUUUCCUCUGCUACACGUGUGAUCAAUUU---GGUUUUC ....((((.....(.(((((((.....(((((((((.....)))))))))((((((.....))))))......................))))))).).)))).......---....... ( -24.10) >DroSec_CAF1 12606 120 - 1 GCACUCAUUAAGCCUUGUGGCAUAAAUAUUUACUUCAGUUUGAGGUGAGUUCAGGGCAUUUUCCUGAUUUAUUACCACAACAUUUCCUCGGCUACACGUGUGAUCAAUUUGUUGGUUUUC ((((......((((((((((.(((((.(((((((((.....)))))))))((((((.....)))))))))))..)))))).........))))....))))((((((....))))))... ( -29.50) >DroSim_CAF1 12901 120 - 1 ACACUCAUUAAGCCUUGUGGCAUAAAUAUUUACUUCAGUUUGAGGUGAGUUCAGGGCAUUUUCCUGAUUUAUUACCACAACAUUUCCUCGGCUACACGUGUGAUCAAUUUGUUGGUUCUC ((((......((((((((((.(((((.(((((((((.....)))))))))((((((.....)))))))))))..)))))).........))))....))))((((((....))))))... ( -29.20) >DroEre_CAF1 10378 117 - 1 ACACUCAUUAAGCCUUGUGGCAUAAAUAUUUACUUCAGCUUGAGGUGAGUUCAGGGCAUUUUCCUGAUUUAUUGCCGCAACAUUUCCUCUGCUACACGUGUGAUCAAUUU---GGGGUUU .........(((((((((((((((((.(((((((((.....)))))))))((((((.....)))))))))).))))))..............(((....)))........---))))))) ( -29.70) >DroYak_CAF1 10414 117 - 1 ACACUCAUUAAGCCUUGUGGCAUAAAUAUUUACUUCAGUUUGAGGUGAGUUCAGGGCAUUUUCCUGAUUUAUUGCCACAACAGUUCCUCUGCUACACGUGUGAUCAAUUU---GGGUUUU .........(((((((((((((((((.(((((((((.....)))))))))((((((.....)))))))))).))))))).(((.....)))...................---)))))). ( -31.20) >consensus ACACUCAUUAAGCCUUGUGGCAUAAAUAUUUACUUCAGUUUGAGGUGAGUUCAGGGCAUUUUCCUGAUUUAUUACCACAACAUUUCCUCUGCUACACGUGUGAUCAAUUU___GGUUUUC ....((((...(...((((((......(((((((((.....)))))))))((((((.....)))))).......................)))))))..))))................. (-23.00 = -23.00 + 0.00)

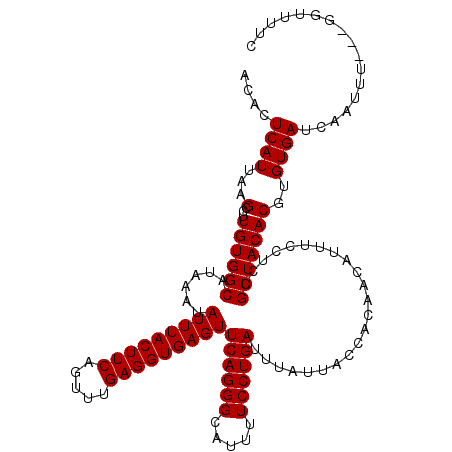
| Location | 15,910,933 – 15,911,053 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -24.14 |
| Energy contribution | -23.62 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15910933 120 + 23771897 UUGUGGUAAUAUAUCAGGAAAAUGCCCUGAACUCACCUCAAACUGAAGUAAAUAUUUAUGCCACAAGACUUAAUGAGUGUUUGAAUAGUAUACUAAAUUUAGUCGGCUACUAAAAGAGGA ((((((((.....(((((.......)))))....((.((.....)).)).........))))))))((((.....(((((.........)))))......))))................ ( -23.70) >DroSec_CAF1 12646 120 + 1 UUGUGGUAAUAAAUCAGGAAAAUGCCCUGAACUCACCUCAAACUGAAGUAAAUAUUUAUGCCACAAGGCUUAAUGAGUGCUUGAAUAGUGUACUAAAUUUAGUCGGCUACUAAAUGAGGG .............(((((.......))))).....(((((...(.(((((..((((...(((....)))..))))..))))).).((((..((((....))))..)))).....))))). ( -27.70) >DroSim_CAF1 12941 120 + 1 UUGUGGUAAUAAAUCAGGAAAAUGCCCUGAACUCACCUCAAACUGAAGUAAAUAUUUAUGCCACAAGGCUUAAUGAGUGUUUGAAUAGUAUACUAAAUUUAGUCGGCUACUAAAAGAGGG .............(((((.......))))).....((((..((((...((((((((((((((....)))...)))))))))))..))))........((((((.....)))))).)))). ( -28.10) >DroEre_CAF1 10415 119 + 1 UUGCGGCAAUAAAUCAGGAAAAUGCCCUGAACUCACCUCAAGCUGAAGUAAAUAUUUAUGCCACAAGGCUUAAUGAGUGUUUGAAUAGUAUACUAAAUUUAGUCGUAUACUAAAAGG-GG .............(((((.......))))).....((((.........((((((((((((((....)))...)))))))))))..((((((((...........))))))))...))-)) ( -27.70) >DroYak_CAF1 10451 119 + 1 UUGUGGCAAUAAAUCAGGAAAAUGCCCUGAACUCACCUCAAACUGAAGUAAAUAUUUAUGCCACAAGGCUUAAUGAGUGUUUGAAUAGUAUACUAAAUUUAGUCGUAUACUAAAAGA-GG .............(((((.......))))).....((((.........((((((((((((((....)))...)))))))))))..((((((((...........))))))))...))-)) ( -28.60) >consensus UUGUGGUAAUAAAUCAGGAAAAUGCCCUGAACUCACCUCAAACUGAAGUAAAUAUUUAUGCCACAAGGCUUAAUGAGUGUUUGAAUAGUAUACUAAAUUUAGUCGGCUACUAAAAGAGGG ((((((((.....(((((.......)))))....((.((.....)).)).........))))))))((((.....(((((.........)))))......))))................ (-24.14 = -23.62 + -0.52)


| Location | 15,910,933 – 15,911,053 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -24.42 |
| Energy contribution | -24.30 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15910933 120 - 23771897 UCCUCUUUUAGUAGCCGACUAAAUUUAGUAUACUAUUCAAACACUCAUUAAGUCUUGUGGCAUAAAUAUUUACUUCAGUUUGAGGUGAGUUCAGGGCAUUUUCCUGAUAUAUUACCACAA ........(((((....((((....)))).)))))....................(((((.......(((((((((.....)))))))))((((((.....)))))).......))))). ( -25.40) >DroSec_CAF1 12646 120 - 1 CCCUCAUUUAGUAGCCGACUAAAUUUAGUACACUAUUCAAGCACUCAUUAAGCCUUGUGGCAUAAAUAUUUACUUCAGUUUGAGGUGAGUUCAGGGCAUUUUCCUGAUUUAUUACCACAA .....(((((((.....)))))))............((((((.........(((....))).(((....))).....))))))(((((..((((((.....))))))....))))).... ( -25.00) >DroSim_CAF1 12941 120 - 1 CCCUCUUUUAGUAGCCGACUAAAUUUAGUAUACUAUUCAAACACUCAUUAAGCCUUGUGGCAUAAAUAUUUACUUCAGUUUGAGGUGAGUUCAGGGCAUUUUCCUGAUUUAUUACCACAA ........(((((....((((....)))).))))).((((((.........(((....))).(((....))).....))))))(((((..((((((.....))))))....))))).... ( -26.60) >DroEre_CAF1 10415 119 - 1 CC-CCUUUUAGUAUACGACUAAAUUUAGUAUACUAUUCAAACACUCAUUAAGCCUUGUGGCAUAAAUAUUUACUUCAGCUUGAGGUGAGUUCAGGGCAUUUUCCUGAUUUAUUGCCGCAA ..-.....((((((((...........))))))))....................(((((((((((.(((((((((.....)))))))))((((((.....)))))))))).))))))). ( -32.80) >DroYak_CAF1 10451 119 - 1 CC-UCUUUUAGUAUACGACUAAAUUUAGUAUACUAUUCAAACACUCAUUAAGCCUUGUGGCAUAAAUAUUUACUUCAGUUUGAGGUGAGUUCAGGGCAUUUUCCUGAUUUAUUGCCACAA ..-.....((((((((...........))))))))....................(((((((((((.(((((((((.....)))))))))((((((.....)))))))))).))))))). ( -33.20) >consensus CCCUCUUUUAGUAGCCGACUAAAUUUAGUAUACUAUUCAAACACUCAUUAAGCCUUGUGGCAUAAAUAUUUACUUCAGUUUGAGGUGAGUUCAGGGCAUUUUCCUGAUUUAUUACCACAA ........(((((....((((....)))).))))).((((((.........(((....))).(((....))).....))))))(((((..((((((.....))))))....))))).... (-24.42 = -24.30 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:22 2006