| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
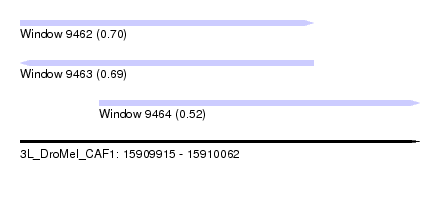
| Location | 15,909,915 – 15,910,062 |
| Length | 147 |
| Max. P | 0.703475 |

| Location | 15,909,915 – 15,910,023 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -20.74 |
| Consensus MFE | -17.84 |
| Energy contribution | -17.92 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15909915 108 + 23771897 GUUUCGUUUCAGUGCGGUUUUUGUUCAUAUUUUCUUGCCAUAUUGUUUUCAGCAUACACUGGCUUUUCCUCAAUUAUGGCGAU-AUGUUCACUUCUAUCCAUCGCCUCU ..........((.(((((...((..((((((.....(((((((((.....(((........)))......))).)))))))))-)))..)).........))))))).. ( -17.80) >DroSec_CAF1 11632 109 + 1 GUUUCGUUUCAGUGCGGGUUUUGUUCAUAUUUUCUUGCCAUAUUGUUUUCAGCAUACACUGGCUUUUCCUCAAUUAUGGCGAUGAUGUUCACUUCUAUCCAUCGCCUGU .........(((.((((((...((..(((((...(((((((((((.....(((........)))......))).)))))))).)))))..))....))))...))))). ( -22.00) >DroSim_CAF1 11897 109 + 1 GUUUCGUUUCAGUGCGGGUUUUGUUCAUAUUUUCUUGCCAUAUUGUUUUCAGCAUACACUGGCUUUUCCUCAAUUAUGGCGAUGAUGUUCACUUCUAUCCAUCGCCUGU .........(((.((((((...((..(((((...(((((((((((.....(((........)))......))).)))))))).)))))..))....))))...))))). ( -22.00) >DroEre_CAF1 9414 109 + 1 GUUUGGCUUCAGAGCGCGUUUUCUUCAUAUUUUCUUGCCAUAUUGCUUUCAGCAUCCACUGGCUUUUCCUCAAUUUUGGCGAUGAUGCUCACUUCUAUCCAUCGCCCGU .....(((....)))(((..................((((...(((.....))).....))))..............((((((((((........))).)))))))))) ( -21.50) >DroYak_CAF1 9437 109 + 1 CUUUUGUUUUAGUGCGGGUUUUCUUCAUAUUCUCUUGCCAUAUUGCUUUCAGCAUCCACUGGCUUUUCCUCAAUUAUGGCGAUGAUGCUCACUUCUAUCCAUCGCCUGU .............((((((.................(((((((((.....(((........)))......))).))))))(((((((........))).)))))))))) ( -20.40) >consensus GUUUCGUUUCAGUGCGGGUUUUGUUCAUAUUUUCUUGCCAUAUUGUUUUCAGCAUACACUGGCUUUUCCUCAAUUAUGGCGAUGAUGUUCACUUCUAUCCAUCGCCUGU ...((((......))))...................((((...(((.....))).....))))..............((((((((((........))).)))))))... (-17.84 = -17.92 + 0.08)



| Location | 15,909,915 – 15,910,023 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15909915 108 - 23771897 AGAGGCGAUGGAUAGAAGUGAACAU-AUCGCCAUAAUUGAGGAAAAGCCAGUGUAUGCUGAAAACAAUAUGGCAAGAAAAUAUGAACAAAAACCGCACUGAAACGAAAC ..(((((...((((...(....).)-)))((((((.(((.((.....))(((....))).....)))))))))....................))).)).......... ( -18.40) >DroSec_CAF1 11632 109 - 1 ACAGGCGAUGGAUAGAAGUGAACAUCAUCGCCAUAAUUGAGGAAAAGCCAGUGUAUGCUGAAAACAAUAUGGCAAGAAAAUAUGAACAAAACCCGCACUGAAACGAAAC .((((((...(((.((.(....).)))))((((((.(((.((.....))(((....))).....)))))))))....................))).)))......... ( -22.10) >DroSim_CAF1 11897 109 - 1 ACAGGCGAUGGAUAGAAGUGAACAUCAUCGCCAUAAUUGAGGAAAAGCCAGUGUAUGCUGAAAACAAUAUGGCAAGAAAAUAUGAACAAAACCCGCACUGAAACGAAAC .((((((...(((.((.(....).)))))((((((.(((.((.....))(((....))).....)))))))))....................))).)))......... ( -22.10) >DroEre_CAF1 9414 109 - 1 ACGGGCGAUGGAUAGAAGUGAGCAUCAUCGCCAAAAUUGAGGAAAAGCCAGUGGAUGCUGAAAGCAAUAUGGCAAGAAAAUAUGAAGAAAACGCGCUCUGAAGCCAAAC ...((((((((.............))))))))........((....((((...(.(((.....))).).))))............(((........)))....)).... ( -21.42) >DroYak_CAF1 9437 109 - 1 ACAGGCGAUGGAUAGAAGUGAGCAUCAUCGCCAUAAUUGAGGAAAAGCCAGUGGAUGCUGAAAGCAAUAUGGCAAGAGAAUAUGAAGAAAACCCGCACUAAAACAAAAG ...((((((((.............))))))))..............((((...(.(((.....))).).)))).................................... ( -19.82) >consensus ACAGGCGAUGGAUAGAAGUGAACAUCAUCGCCAUAAUUGAGGAAAAGCCAGUGUAUGCUGAAAACAAUAUGGCAAGAAAAUAUGAACAAAACCCGCACUGAAACGAAAC .((((((...(((.((.(....).)))))((((((.(((......(((........))).....)))))))))....................))).)))......... (-18.10 = -18.74 + 0.64)



| Location | 15,909,944 – 15,910,062 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.69 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -21.22 |
| Energy contribution | -21.18 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15909944 118 + 23771897 UUUUCUUGCCAUAUUGUUUUCAGCAUACACUGGCUUUUCCUCAAUUAUGGCGAU-AUGUUCACUUCUAUCCAUCGCCUCUUAUUGUUAUUAUCGGC-AGGGGAAUGGGAUAAACUAGACC ((..((((((.........((((......))))........((((...((((((-..((........))..))))))....))))........)))-)))..))(((......))).... ( -25.20) >DroSec_CAF1 11661 119 + 1 UUUUCUUGCCAUAUUGUUUUCAGCAUACACUGGCUUUUCCUCAAUUAUGGCGAUGAUGUUCACUUCUAUCCAUCGCCUGUUAGUGUUAUUAUCGGC-AGGGGAAUGGGGUAAAAUGGACC ...(((((((((((((.....(((........)))......))).)))))))).)).(((((.((.(((((.((.(((((..(((....)))..))-))).))...))))).))))))). ( -35.50) >DroSim_CAF1 11926 119 + 1 UUUUCUUGCCAUAUUGUUUUCAGCAUACACUGGCUUUUCCUCAAUUAUGGCGAUGAUGUUCACUUCUAUCCAUCGCCUGUUAUUGUUAUUAUCGGC-AGGGGAUUGGGGUAAACUGGACC ...(((((((((((((.....(((........)))......))).)))))))).)).(((((.((.((((((((.((((((..((....))..)))-))).)))..))))))).))))). ( -32.30) >DroEre_CAF1 9443 108 + 1 UUUUCUUGCCAUAUUGCUUUCAGCAUCCACUGGCUUUUCCUCAAUUUUGGCGAUGAUGCUCACUUCUAUCCAUCGCCCGUAAUUGUUAUUAUCGGA-AGGGGACUGG-GU---------- .......((((...(((.....))).....))))...(((((......((((((((((........))).)))))))((((((....)))).))..-.)))))....-..---------- ( -26.10) >DroYak_CAF1 9466 119 + 1 UUCUCUUGCCAUAUUGCUUUCAGCAUCCACUGGCUUUUCCUCAAUUAUGGCGAUGAUGCUCACUUCUAUCCAUCGCCUGUAAUUGUUAUUAUCGGAAAGGGGAAUGG-GUAAAAUAGACC .(((.(((((.((((.(..((((......))))(((((((.(((((((((((((((((........))).))))))).)))))))........)))))))).)))))-))))...))).. ( -31.10) >consensus UUUUCUUGCCAUAUUGUUUUCAGCAUACACUGGCUUUUCCUCAAUUAUGGCGAUGAUGUUCACUUCUAUCCAUCGCCUGUUAUUGUUAUUAUCGGC_AGGGGAAUGGGGUAAAAUAGACC .......((((...(((.....))).....))))...(((((......((((((((((........))).)))))))......((.......))....)))))................. (-21.22 = -21.18 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:16 2006