
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,908,613 – 15,908,886 |
| Length | 273 |
| Max. P | 0.924891 |

| Location | 15,908,613 – 15,908,726 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -15.74 |
| Energy contribution | -16.34 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15908613 113 + 23771897 GCUAAAAGCACAAAAGACAGU-------CCCGAAAGGAAUCCCUGCCACACACGCCGUCAGACACUCGGCGAUUUCAAUUUGAUUUCAAUUCGAUUUCAAUUUGAUUUCAAUUGGCAAAC ((.....))............-------.((....))......(((((.....((((.........))))((..((((.((((..((.....))..)))).))))..))...)))))... ( -29.70) >DroSec_CAF1 8089 113 + 1 GCUAAAAGCACAAAAGACAGC-------CCCGAAAGGAAUUCCUGCCACACACGCCGUCUGACACUCGACGAUUUCAAUUUGAUUUCAAUACGAAUUCAAUUUGAUUUCAAUUGGCAAAC ((.....))............-------.((....))......(((((........(((........)))((..((((.((((.(((.....))).)))).))))..))...)))))... ( -26.20) >DroSim_CAF1 10594 113 + 1 GCUAAAAGCUCAAAAGACAGC-------CCCGAAAGGAAUUCCUGCCACACACGCCGUCUGACACUCGGCGAUUUCAAUUCGAUUUCAAUUCGAUUUCAAUUUGAUUUCAAUUGGCAAAC .......(((........)))-------.((....))......(((((.....((((.........))))((..((((...((..((.....))..))...))))..))...)))))... ( -25.70) >DroEre_CAF1 8170 90 + 1 UCUAAAAGCACAAAA-ACAGC-------CCCAAAAGGAACCACUGCCACACACGCCGUCUGACACUCGGCGAUUUCAAUUCGA----------------------UUUCAAUUGGCAAAA ...............-.....-------.((....))......(((((.....((((.........))))((..((.....))----------------------..))...)))))... ( -15.80) >DroYak_CAF1 8150 98 + 1 GCUAAAAGCACAAAAGACAGCCAAAAUACCCAAAAGGAACGCCUGCCACACACGCCGUCUGACACUCGGCGAUUUCAAUUCGA----------------------UUUCAAUUGGCAAAC ((.....))..........(((((..........(((....)))........(((((.........)))))............----------------------......))))).... ( -18.60) >consensus GCUAAAAGCACAAAAGACAGC_______CCCGAAAGGAAUCCCUGCCACACACGCCGUCUGACACUCGGCGAUUUCAAUUCGAUUUCAAU_CGA_UUCAAUUUGAUUUCAAUUGGCAAAC .............................((....))......(((((....(((((.........)))))..........((..((((............))))..))...)))))... (-15.74 = -16.34 + 0.60)

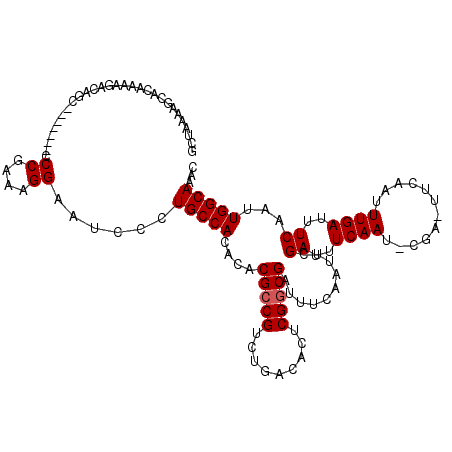

| Location | 15,908,646 – 15,908,766 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.56 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15908646 120 + 23771897 CCCUGCCACACACGCCGUCAGACACUCGGCGAUUUCAAUUUGAUUUCAAUUCGAUUUCAAUUUGAUUUCAAUUGGCAAACGGCAUCGGACUCUUUUGUUUUGGCCAAUUUUAUGUCCCAU ...(((((.....((((.........))))((..((((.((((..((.....))..)))).))))..))...)))))...(((...((((......))))..)))............... ( -31.60) >DroSec_CAF1 8122 120 + 1 UCCUGCCACACACGCCGUCUGACACUCGACGAUUUCAAUUUGAUUUCAAUACGAAUUCAAUUUGAUUUCAAUUGGCAAACGGCAUCGGACUCUUGUGUUUUGCCCAAUUUUAUGUCCCAU ...(((((........(((........)))((..((((.((((.(((.....))).)))).))))..))...)))))...((((..(.((....)).)..))))................ ( -27.90) >DroSim_CAF1 10627 120 + 1 UCCUGCCACACACGCCGUCUGACACUCGGCGAUUUCAAUUCGAUUUCAAUUCGAUUUCAAUUUGAUUUCAAUUGGCAAACGGCAUCGGACUCUUGUGUUUUGGCCAAUUUUAUGUCCCAU ...(((((.....((((.........))))((..((((...((..((.....))..))...))))..))...)))))...(((...((((......))))..)))............... ( -27.80) >DroEre_CAF1 8202 98 + 1 CACUGCCACACACGCCGUCUGACACUCGGCGAUUUCAAUUCGA----------------------UUUCAAUUGGCAAAAGGCAACGGGCUCUUGUCUCUUGGCCAACUUUAUGUCCCAA ...(((((.....((((.........))))((..((.....))----------------------..))...)))))...(((...((((....))))....)))............... ( -24.00) >DroYak_CAF1 8190 98 + 1 GCCUGCCACACACGCCGUCUGACACUCGGCGAUUUCAAUUCGA----------------------UUUCAAUUGGCAAACGGCAACGGGCUCUUGUGUUUUGGCCAGUUUUAUGUCCCAU ((((((((.....((((.........))))((..((.....))----------------------..))...)))))..((....)))))..........(((.((......))..))). ( -24.80) >consensus CCCUGCCACACACGCCGUCUGACACUCGGCGAUUUCAAUUCGAUUUCAAU_CGA_UUCAAUUUGAUUUCAAUUGGCAAACGGCAUCGGACUCUUGUGUUUUGGCCAAUUUUAUGUCCCAU ...((((.....(((((.........)))))...((((((.((..((((............))))..)))))))).....))))..((((.......................))))... (-19.20 = -19.56 + 0.36)

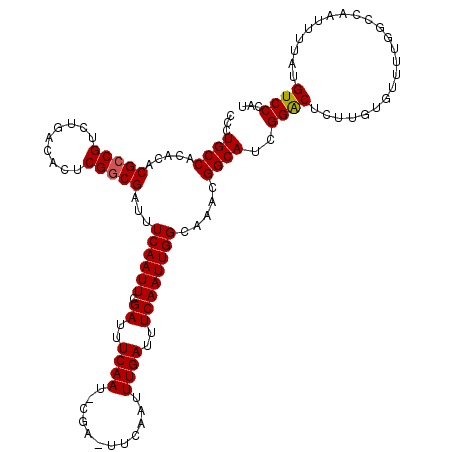
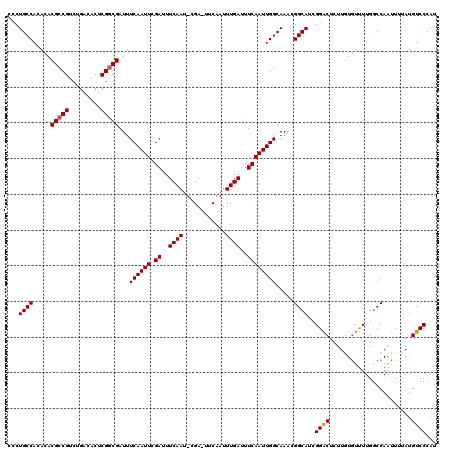
| Location | 15,908,766 – 15,908,886 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -24.12 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15908766 120 - 23771897 ACUUUUUAACGGAUCGUGAUGAGGUAUAUUUUUGAGCUGGCAAUCCUUUUAGUCUGCCCAUUGAUUGAUGGAUUUGGCAAAGAUAAAUAUUUACGCACGAUUCUGGGAAACACAAUUCAG ..........((((((((...((((((...(((..((..(..((((..((((((........)))))).)))))..))..)))...))))))...))))))))(((....).))...... ( -29.90) >DroSec_CAF1 8242 120 - 1 AUUUUUCAAUGGAUCGUGCUAAGGUAUAUUUUUGAGCUAACAAUCCUUUUAGGCUGCCCAUUGAUUGAUGGAUUUGGCAAAGAUAAAUAUUUACGCACGAUUCUGGGAAACACAAUUCAG .(((..((..(((((((((((((...((((((((((((.............))))..(((((....)))))......))))))))....)))).)))))))))))..))).......... ( -30.02) >DroSim_CAF1 10747 120 - 1 ACUUUUCAAUAGAUCGUGCUGAGGUAUAUUUUUGAGCUAACAAUCCUUUUAGUCUGCCCAUUGAUUGAUGGAUUUGGCAAAGAUAAAUAUUUACGCACGAUUCUGGGAAACACAAUUCAG ..((..((...((((((((..((((((...(((..(((((..((((..((((((........)))))).)))))))))..)))...))))))..)))))))).))..))........... ( -30.30) >DroEre_CAF1 8300 118 - 1 ACUUUUCAAUGGAUCGUGGCGCGGUA-AUUUUUGAACUGGCAAUCCUUUUAGUCUGCCCAUUGAUUGAUGGAU-UUGCAGAGAUAAAUAUUUACGCACGGUUCUGGGAAACACAAUUCAG ...((((...((((((((.((((((.-........))))(((((((..((((((........)))))).))).-))))...............))))))))))...)))).......... ( -28.40) >DroYak_CAF1 8288 120 - 1 ACUUUUCAAUGGAUCGUGGUGCGGUAUAUUUUUGAGCUGGCAAUCCUUUUAGUCUGCCCAUUGAUUGAUGGAUUUUGCACAGAUAAAUAUUUACGCACGGUUCUGGGAAAUACAAUUCAG ...((((...(((((...(((((..((((((.....((((((((((..((((((........)))))).))))..))).)))..))))))...))))))))))...)))).......... ( -31.70) >consensus ACUUUUCAAUGGAUCGUGCUGAGGUAUAUUUUUGAGCUGGCAAUCCUUUUAGUCUGCCCAUUGAUUGAUGGAUUUGGCAAAGAUAAAUAUUUACGCACGAUUCUGGGAAACACAAUUCAG ..(((((...((((((((...(((((((((((((.......(((((..((((((........)))))).)))))...))))))))..)))))...))))))))..))))).......... (-24.12 = -25.00 + 0.88)


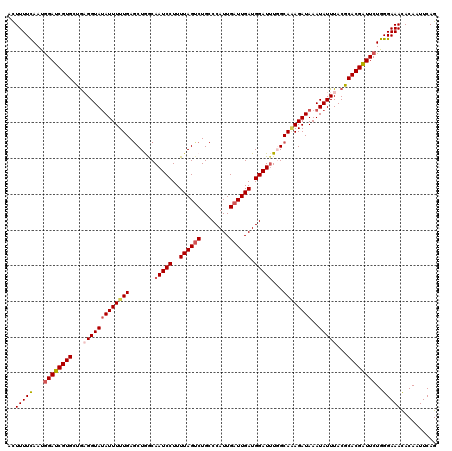
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:10 2006