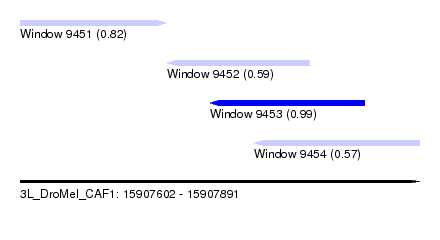
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,907,602 – 15,907,891 |
| Length | 289 |
| Max. P | 0.987756 |

| Location | 15,907,602 – 15,907,708 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.89 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15907602 106 + 23771897 CUUUCUAGAAGGGUUUCUUUACA-AUUCUUCCUGGUUUAUAGC---------CUU----GCACGAAUGGGGAAAGAUCUGUACUGCGGAAAUUACCUAUCCGCCCACAGAACACCUCUCA .......((..(((((((.((((-..(((((((.(((..(.((---------...----)))..))).))).))))..))))..(((((.........)))))....)))).)))..)). ( -27.30) >DroSec_CAF1 7094 106 + 1 CUUUCUAGAAGGCUUUCUUUGCA-AUUCUUCCUGGUUUAUAGC---------CUU----GCAUGAAUGUGUAAAGAUCUGUACUGCGGAAAUUACCUAUCCGCCCACCGAACACCUCUCA .......(((((...((((((((-((((.....(((.....))---------)..----....)))).))))))))((.((...(((((.........)))))..)).))...))).)). ( -23.10) >DroSim_CAF1 9599 105 + 1 CUUUCUAGAAGGGUUUCUUUGCA-AUUCUUCCUGGUUUAUAGC----------UU----GCAUGAAUGUGUAAAGAUCUGUACUGCGGAAAUUACCUAUCCGCCCACCGAACACCUCUCA .......((..(((.((((((((-((((.....(((.....))----------).----....)))).))))))))((.((...(((((.........)))))..)).))..)))..)). ( -24.60) >DroEre_CAF1 7173 111 + 1 CUUUCUAGAAGGGUUG--UUCCCAUUUCUUCCUGGUU-----CACUUACAGCAAUAUAGACACGAAAAUGGAAAUAUCUGUAUGGCGGAAAUUACCAAUCCGCCCACAGAA--CCUCUCA .......((..(((((--((.((((((..((.((...-----..................)).)))))))).))))(((((..((((((.........)))))).))))))--))..)). ( -26.50) >DroYak_CAF1 7111 117 + 1 AUUUCUAGAAGUGUUUCUUUCCAAUUUCUUCCGGGUU-AUACCAUAUACAGCAAUAUUGACGCGAAAUGGGAAAGAUCUGUACUGCGGAAAUUACCAAUCCGCCCACAGAA--CCUCUCA ......(((.((...(((((((.(((((....((...-...)).......((.........))))))).)))))))(((((...(((((.........)))))..))))))--).))).. ( -29.20) >consensus CUUUCUAGAAGGGUUUCUUUCCA_AUUCUUCCUGGUUUAUAGC_________AUU____GCACGAAUGUGGAAAGAUCUGUACUGCGGAAAUUACCUAUCCGCCCACAGAACACCUCUCA .......(((((...(((((((..((((...................................))))..)))))))(((((...(((((.........)))))..)))))...))).)). (-16.29 = -16.89 + 0.60)



| Location | 15,907,708 – 15,907,811 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.57 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -22.07 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15907708 103 - 23771897 GAGGCAGGUGGGCUAAUUUCUAUAUGCCAUGUGGUAAAUGGUAAACAUCUGCUUCGAAAGUGUCAUAAAUCG-----------------AUGUCGCAAAUGAAAGGGUGAUGGGGCAAGG ((((((((((.((((.((.((((((...)))))).)).))))...))))))))))....((.((((...((.-----------------...(((....)))..))...)))).)).... ( -28.90) >DroSec_CAF1 7200 120 - 1 GGGGCAGGUGGGCUAAUUUCUAGAUGCCAUGUGGUAAAUGGUAAACAUCUGCUUCGAAAGUGUCAUAAAUCGCAUAAGUUAUGACUGGGAUGUCGCAAGUGGAAGGGUGAUGGGGCAAGG ((((((((((..(((.....))).((((((.......))))))..)))))))))).....((((....(((((...(.((.((((......)))).)).)......)))))..))))... ( -33.60) >DroSim_CAF1 9704 120 - 1 GGGGCAGGUGGGCUAAUUUCUAGAUGCCAUGUGGUAAAUGGUAAACAUCUGCUUCGAAAGUGUCAUAAAUCGCCUAAGUUAUGACUGGGAUGUCGCAAAUGGAAGGGUGAUGGGGCAAGG ((((((((((..(((.....))).((((((.......))))))..)))))))))).....((((....(((((((..(((.((((......))))..)))....)))))))..))))... ( -37.20) >DroEre_CAF1 7284 107 - 1 GGGGCAGGUGGGUUAAUUUCUAUUUGCUAUGUGGUAAAUGGUAAACAUCUGCUUCGAAAGUGUCAUAAAUCGCUUAGGUUAUGACUGGGAUGUCGCAAGUGGAAG-------------GG ((((((((((..(......(((((((((....))))))))).)..))))))))))......(((((((.((.....)))))))))((.(....).))........-------------.. ( -29.60) >DroYak_CAF1 7228 119 - 1 GGGGCAGGUGGGCUAAUUUCUAUAUGCUAUGUGGUAAAUGGUAAACAUCUGCUUCGAAAGUGUCAUAAAUCGCUUAAGUUAUGACUGGGAUGUCGCAAAUGGAAGGGC-AUGGGGCGGGG ((((((((((.((((.((.((((((...)))))).)).))))...))))))))))......(((((((..........))))))).......((((..(((......)-))...)))).. ( -32.50) >consensus GGGGCAGGUGGGCUAAUUUCUAUAUGCCAUGUGGUAAAUGGUAAACAUCUGCUUCGAAAGUGUCAUAAAUCGCUUAAGUUAUGACUGGGAUGUCGCAAAUGGAAGGGUGAUGGGGCAAGG ((((((((((.........((((.((((....)))).))))....))))))))))....(((.(((......(.........)......))).)))........................ (-22.07 = -22.12 + 0.05)

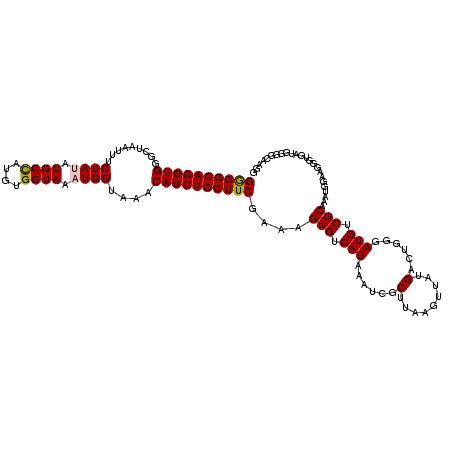
| Location | 15,907,739 – 15,907,851 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -26.98 |
| Energy contribution | -26.98 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15907739 112 - 23771897 UGUAAAAACUUGACAAAUGUGAAAACAUCAUCGACAGGCAGAGGCAGGUGGGCUAAUUUCUAUAUGCCAUGUGGUAAAUGGUAAACAUCUGCUUCGAAAGUGUCAUAAAUCG-------- ..........(((((.((((....))))............((((((((((.((((.((.((((((...)))))).)).))))...)))))))))).....))))).......-------- ( -30.10) >DroSec_CAF1 7240 120 - 1 UGUAAAAACUUGACAAAUGUGAAAACAUCAUCGACAGGCAGGGGCAGGUGGGCUAAUUUCUAGAUGCCAUGUGGUAAAUGGUAAACAUCUGCUUCGAAAGUGUCAUAAAUCGCAUAAGUU ......((((((....((((....))))....((((....((((((((((..(((.....))).((((((.......))))))..)))))))))).....))))..........)))))) ( -31.30) >DroSim_CAF1 9744 120 - 1 UGUAAAAACUUGACAAAUGUGAAAACAUCAUCGACAGGCAGGGGCAGGUGGGCUAAUUUCUAGAUGCCAUGUGGUAAAUGGUAAACAUCUGCUUCGAAAGUGUCAUAAAUCGCCUAAGUU ......(((((((...((((....))))..))...((((.((((((((((..(((.....))).((((((.......))))))..))))))))))((.....)).......))))))))) ( -32.10) >DroEre_CAF1 7311 119 - 1 UGUAA-AACUUGACAAAUGUGAAAACAUCACCGACAGGCAGGGGCAGGUGGGUUAAUUUCUAUUUGCUAUGUGGUAAAUGGUAAACAUCUGCUUCGAAAGUGUCAUAAAUCGCUUAGGUU .....-((((((((..((((....))))....((((....((((((((((..(......(((((((((....))))))))).)..)))))))))).....)))).......).))))))) ( -32.80) >DroYak_CAF1 7267 120 - 1 UGUAAAAACUUGACAAAUGUGAAAACAUCAUCGACAGGCAGGGGCAGGUGGGCUAAUUUCUAUAUGCUAUGUGGUAAAUGGUAAACAUCUGCUUCGAAAGUGUCAUAAAUCGCUUAAGUU ......((((((((..((((....))))....((((....((((((((((.((((.((.((((((...)))))).)).))))...)))))))))).....)))).......).))))))) ( -32.40) >consensus UGUAAAAACUUGACAAAUGUGAAAACAUCAUCGACAGGCAGGGGCAGGUGGGCUAAUUUCUAUAUGCCAUGUGGUAAAUGGUAAACAUCUGCUUCGAAAGUGUCAUAAAUCGCUUAAGUU ..........(((((.((((....))))............((((((((((.........((((.((((....)))).))))....)))))))))).....)))))............... (-26.98 = -26.98 + 0.00)

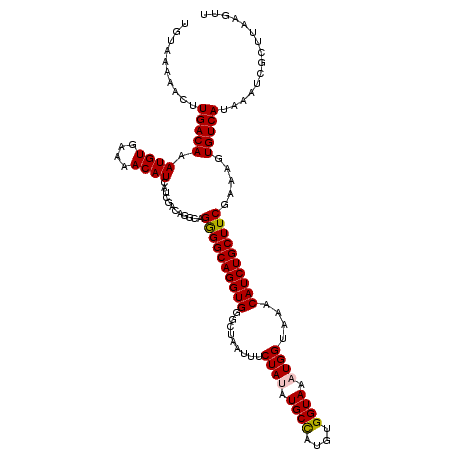
| Location | 15,907,771 – 15,907,891 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -31.20 |
| Energy contribution | -31.04 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15907771 120 - 23771897 GAUCACGCCCCCACCACUUGGCGGUCUUGUAAAGCUGCAGUGUAAAAACUUGACAAAUGUGAAAACAUCAUCGACAGGCAGAGGCAGGUGGGCUAAUUUCUAUAUGCCAUGUGGUAAAUG .((((((((.....((((..(((((........))))))))).......((((...((((....))))..))))..)))...((((.(((((......))))).))))..)))))..... ( -32.00) >DroSec_CAF1 7280 120 - 1 GAUCACGCCCCCACCACUUGGCGGUCUUGUAAAGCUGCGAUGUAAAAACUUGACAAAUGUGAAAACAUCAUCGACAGGCAGGGGCAGGUGGGCUAAUUUCUAGAUGCCAUGUGGUAAAUG .((((((((((..((((.(.(((((........))))).).))......((((...((((....))))..))))..))..))))).((((..(((.....))).))))..)))))..... ( -35.10) >DroSim_CAF1 9784 120 - 1 GAUCACGCCCCCACCACUUGGCGGUCUUGUAAAGCUGCGAUGUAAAAACUUGACAAAUGUGAAAACAUCAUCGACAGGCAGGGGCAGGUGGGCUAAUUUCUAGAUGCCAUGUGGUAAAUG .((((((((((..((((.(.(((((........))))).).))......((((...((((....))))..))))..))..))))).((((..(((.....))).))))..)))))..... ( -35.10) >DroEre_CAF1 7351 119 - 1 GACCACGCCCCCACCACUUUGCGGUCUUGUAAGGCUGCAGUGUAA-AACUUGACAAAUGUGAAAACAUCACCGACAGGCAGGGGCAGGUGGGUUAAUUUCUAUUUGCUAUGUGGUAAAUG .((((((((((..((((.(((((((((....))))))))).))..-..........((((....))))........))..))))).((..(((........)))..))..)))))..... ( -40.60) >DroYak_CAF1 7307 120 - 1 GAUCACGCCCCCACCACUUUGCGGUCUUGUAAAGCUGCAGUGUAAAAACUUGACAAAUGUGAAAACAUCAUCGACAGGCAGGGGCAGGUGGGCUAAUUUCUAUAUGCUAUGUGGUAAAUG .((((((((((..((((.(((((((........))))))).))......((((...((((....))))..))))..))..)))))..(((((......))))).......)))))..... ( -33.70) >consensus GAUCACGCCCCCACCACUUGGCGGUCUUGUAAAGCUGCAGUGUAAAAACUUGACAAAUGUGAAAACAUCAUCGACAGGCAGGGGCAGGUGGGCUAAUUUCUAUAUGCCAUGUGGUAAAUG .((((((((((..((.....(((((........)))))...........((((...((((....))))..))))..))..))))).....(((............)))..)))))..... (-31.20 = -31.04 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:06 2006