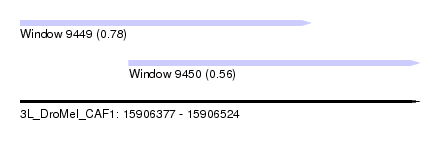
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,906,377 – 15,906,524 |
| Length | 147 |
| Max. P | 0.784249 |

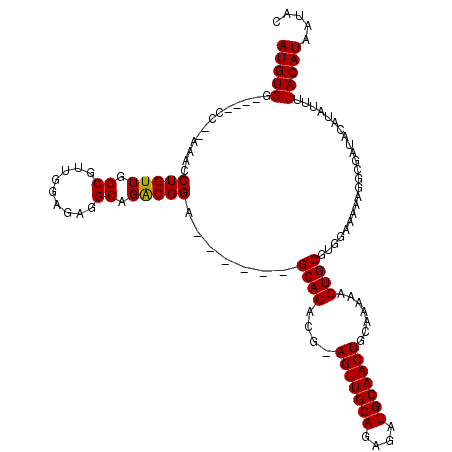
| Location | 15,906,377 – 15,906,484 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.98 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.24 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15906377 107 + 23771897 GACCAAGUCGAUAGGCGGCCACAAUUAAAGUCUGGCCGAGAUGUGG----CC--AAACCUGUUGGCGUUCGAGAGGCAGACGGA------GCACACG-AGUUGCAGAGAUGCAACUGCAA ..((..(((((((((.(((((((.......(((.....))))))))----))--...)))))))))((.(....)))....)).------(((....-.((((((....))))))))).. ( -36.31) >DroSec_CAF1 5774 108 + 1 GACCAAGUCGAUAGGCGGCCACAAUUAAAGUCUGGCCGAGAUGUGG----CC--AAACCUGUUGGCAAUGGAGAAGCAGGCGGA------GCACACAAAGUUGCACAUAUGCAACUGCAA ..(((.(((((((((.(((((((.......(((.....))))))))----))--...)))))))))..)))....((..((...------))......(((((((....))))))))).. ( -35.61) >DroSim_CAF1 8245 108 + 1 GACCAAGUCGAUAGGCGGCCACAAUUAAAGUCUGGCCGAGAUGUGG----CC--AAACCUGUUGGCGUUGGAGAGGCAGACGGA------GCACACAAAGUUGCACAUAUGCAACUGCAA ..(((((((((((((.(((((((.......(((.....))))))))----))--...))))))))).))))....((....(..------...)....(((((((....))))))))).. ( -37.21) >DroEre_CAF1 5963 107 + 1 GACCAAGUCGGCAGUCGGCCACAAUUAAAGUCUGGCCGAGAUGUGG----GC--AAACCUGUUGGCGAUGGAGAGGCAGACGGA------GCACACG-AGUUGCAGAGAUGCAACUGGAA ..((..(((.(((.(((((((...........)))))))..))).)----))--....(((((..(......).)))))..)).------.....(.-(((((((....))))))).).. ( -37.10) >DroYak_CAF1 5874 119 + 1 GACCAAGUCGAUAGUCGGCCACAAUUAAAGUCUGGCCGAGAUGUGGCUGUGCAAAAACCUGUUGGCGUUGGAGAGGCAGACGGAGACGGCGCACAUG-AGUUGCAGAGAUGCAACUGGAA ..((.(((((....(((((((...........)))))))....)))))((((......(((((..(......).))))).((....))..))))...-(((((((....))))))))).. ( -41.40) >consensus GACCAAGUCGAUAGGCGGCCACAAUUAAAGUCUGGCCGAGAUGUGG____CC__AAACCUGUUGGCGUUGGAGAGGCAGACGGA______GCACACG_AGUUGCAGAGAUGCAACUGCAA ..(((.(((......((((((...........)))))).))).)))............(((((.((.........)).)))))...............(((((((....))))))).... (-27.40 = -27.24 + -0.16)

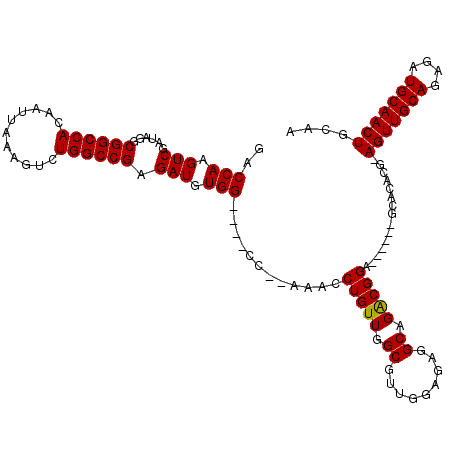
| Location | 15,906,417 – 15,906,524 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.69 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -21.86 |
| Energy contribution | -21.70 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15906417 107 + 23771897 AUGUGG----CC--AAACCUGUUGGCGUUCGAGAGGCAGACGGA------GCACACG-AGUUGCAGAGAUGCAACUGCAAAAAGUGCGUGGAAAAAGGCGAUACAUAUUUUACAUAAUAC .(((.(----((--....(((((.((.((...)).)).))))).------...(((.-(((((((....)))))))(((.....))))))......))).)))................. ( -28.80) >DroSec_CAF1 5814 108 + 1 AUGUGG----CC--AAACCUGUUGGCAAUGGAGAAGCAGGCGGA------GCACACAAAGUUGCACAUAUGCAACUGCAAAAAGUGCGUGGAAAAAGGCGAUACAUAUUUUACAUAAUAC .(((.(----((--...((((((..(....)...))))))....------((((....(((((((....))))))).......)))).........))).)))................. ( -27.40) >DroSim_CAF1 8285 108 + 1 AUGUGG----CC--AAACCUGUUGGCGUUGGAGAGGCAGACGGA------GCACACAAAGUUGCACAUAUGCAACUGCAAAAAGUGCGUGGAAAAAGGCGAUACAUAUUUUACAUAAUAC .(((.(----((--....(((((.((.((...)).)).))))).------((((....(((((((....))))))).......)))).........))).)))................. ( -27.70) >DroEre_CAF1 6003 107 + 1 AUGUGG----GC--AAACCUGUUGGCGAUGGAGAGGCAGACGGA------GCACACG-AGUUGCAGAGAUGCAACUGGAAAAAGUGCGUGGAAAAAGGCGAAACAUAUUUUACAUAAUAC ((((..----((--....(((((.((.........)).))))).------((((.(.-(((((((....))))))).).....))))..........))...)))).............. ( -25.00) >DroYak_CAF1 5914 119 + 1 AUGUGGCUGUGCAAAAACCUGUUGGCGUUGGAGAGGCAGACGGAGACGGCGCACAUG-AGUUGCAGAGAUGCAACUGGAAAAAGUGCGUGGAAAAAGGCGAAACAUAUUUUACAUAAUAC (((((....(((......(((((.((.((...)).)).)))))...(.((((((.(.-(((((((....))))))).).....)))))).)......)))...)))))............ ( -31.90) >consensus AUGUGG____CC__AAACCUGUUGGCGUUGGAGAGGCAGACGGA______GCACACG_AGUUGCAGAGAUGCAACUGCAAAAAGUGCGUGGAAAAAGGCGAUACAUAUUUUACAUAAUAC (((((.............(((((.((.........)).))))).......((((....(((((((....))))))).......)))).......................)))))..... (-21.86 = -21.70 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:02 2006