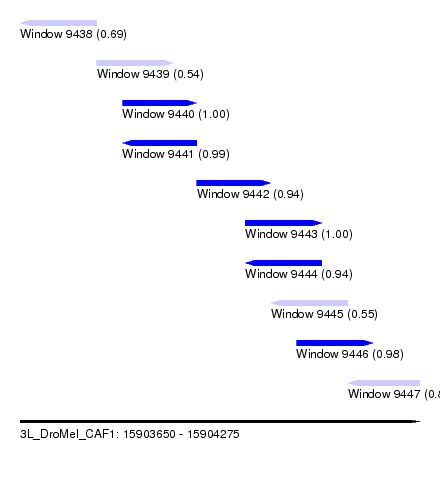
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,903,650 – 15,904,275 |
| Length | 625 |
| Max. P | 0.999020 |

| Location | 15,903,650 – 15,903,770 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -22.16 |
| Energy contribution | -24.36 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15903650 120 - 23771897 UUUCGUAGCCUCAAAACGUGCUCGACAGUGAAAACAGUAAUUAUGCAAACAAUUGAGGUUCGAAAAGUGGUUCUAAAGAUAGCGCUCAUUAAAUGGGAAUAUAAUAUAGGGAACUGGAAG (((((.((((((((....(((...((.((....)).))......))).....)))))))))))))...((((((....(((...(((((...)))))..))).......))))))..... ( -28.50) >DroSec_CAF1 3065 120 - 1 UUUCGUAGCCUCAAAACGUGCUCGACAGUGAAAACAGUAAUUAUGCAAACAGUUGAGGUUCGAAAAGUGGUGCUAAAGAUAGCGCUCAUUAAAUGGGAAUAUAAUAUAGGGAACUGCAAG (((((.((((((((....(((...((.((....)).))......))).....)))))))))))))((((((((((....)))))).))))................(((....))).... ( -31.80) >DroSim_CAF1 5268 120 - 1 UUUCGUAGCCUCAAAACGUGCUCGACAGUGAAAACAGUAAUUAUGCAAACAGUUGAGGUUCGAAAAGUGGUUCCAAAAAUAGCGCUCAUUAAAUGGCAAUAUAAUAUAGGGAACUGCAAG (((((.((((((((....(((...((.((....)).))......))).....))))))))))))).(..(((((....(((..(((........)))..))).......)))))..)... ( -33.20) >DroEre_CAF1 2993 116 - 1 ----CUAGCCUCAAAACGUGCUCGACAGUGAAAACAGUAAUUAUGCAAACAGCUGAGGUUCGAAAAGUGGUUCCAAAGAUAGCGCUCAUUAAAUGGGAAUAUACUAUAGGGCACAGGAAG ----(.(((((((.....(((...((.((....)).))......)))......))))))).)....(((..(((..(((((...(((((...)))))..))).))...))))))...... ( -20.50) >DroYak_CAF1 3073 117 - 1 UUUCGUAGCCUUAAAACGUGCUCGACAGUGAAAACAGUAAUUAUGCAAACAGUUG-GGUUCGAAAAGUGGUUCUAAAGAUAGCGCUCAUUAAAUGGCAAUAUACUAUAGGGAACCGUA-- (((((.(((((.......(((...((.((....)).))......))).......)-)))))))))...((((((..(((((..(((........)))..))).))....))))))...-- ( -26.04) >consensus UUUCGUAGCCUCAAAACGUGCUCGACAGUGAAAACAGUAAUUAUGCAAACAGUUGAGGUUCGAAAAGUGGUUCUAAAGAUAGCGCUCAUUAAAUGGGAAUAUAAUAUAGGGAACUGCAAG (((((.((((((((....(((...((.((....)).))......))).....))))))))))))).((((((((....(((...(((((...)))))..))).......))))))))... (-22.16 = -24.36 + 2.20)


| Location | 15,903,770 – 15,903,890 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -27.59 |
| Consensus MFE | -23.75 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15903770 120 + 23771897 GCCAGCUCGCUUCUACCACUACCAUUACUACUUAUACUACCAGGUAAUAUGACCAACAUGGGUGUUGGCUUACACGCCCGGCCGAGAUUUUCUCAACCUUGAGUUUUUAAUUGGGCUAAC ((((((.(..............(((...(((((........)))))..))).((.....))).))))))......((((((..((((....((((....)))).))))..)))))).... ( -27.10) >DroSec_CAF1 3185 120 + 1 GCCAGCUCGCUUCUACCACUACCACUACUACUUAUACCACCAGGUAAUAUGACCAACAUGGGUGUUGGCUUACACGCCCGCCCGAGAUUUUCUCAACCUUGAGUUUUUAAUUGGGCUAAC ((((((.(..........................((((....))))......((.....))).))))))..........((((((......((((....)))).......)))))).... ( -28.42) >DroSim_CAF1 5388 120 + 1 GCCAGCUCGCUUCUACCACUACCACUACUACCUAUACCACCAGGUAAUAUGACCAACAUGGGUGUUGGCUUACACGCCCGCCCGAGAUUUUCUCAACCUUGAGUUUUUAAUUGGGCUAAC ((((((.(....................(((((........)))))......((.....))).))))))..........((((((......((((....)))).......)))))).... ( -29.12) >DroYak_CAF1 3190 113 + 1 GCCAGCUCGAUUCUACCACUACUAGCACCAC---UACCAC----UAAUAUGACCAACAUGGGUGUUGGCUUACACGCCCGCCCGAGAUUUUCUCAACCUUGAGUUUUUAAUUGGGCUAAC ...(((((((((.....(((...((......---......----...............(((((..(((......))))))))(((.....)))...))..)))....)))))))))... ( -25.70) >consensus GCCAGCUCGCUUCUACCACUACCACUACUACUUAUACCACCAGGUAAUAUGACCAACAUGGGUGUUGGCUUACACGCCCGCCCGAGAUUUUCUCAACCUUGAGUUUUUAAUUGGGCUAAC ((((((.(..........................((((....))))......((.....))).))))))......(((((...((((....((((....)))).))))...))))).... (-23.75 = -24.12 + 0.38)

| Location | 15,903,810 – 15,903,926 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.20 |
| Mean single sequence MFE | -47.13 |
| Consensus MFE | -38.60 |
| Energy contribution | -40.80 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15903810 116 + 23771897 CAGGUAAUAUGACCAACAUGGGUGUUGGCUUACACGCCCGGCCGAGAUUUUCUCAACCUUGAGUUUUUAAUUGGGCUAACGCCCGACUCGCUGGGC----ACUCGGUGGGCCAACGAGUG ..(((......)))..(((...(((((((((((..(((((((.(((.....((((....)))).......((((((....))))))))))))))))----.....))))))))))).))) ( -51.00) >DroSec_CAF1 3225 116 + 1 CAGGUAAUAUGACCAACAUGGGUGUUGGCUUACACGCCCGCCCGAGAUUUUCUCAACCUUGAGUUUUUAAUUGGGCUAACGCCCGACUCGCUGGGC----ACUCGGUGGGCCAACGAGUG ..(((......)))..(((...(((((((((((.((...(((((((.....)))......(((((.......((((....)))))))))...))))----...))))))))))))).))) ( -46.21) >DroSim_CAF1 5428 116 + 1 CAGGUAAUAUGACCAACAUGGGUGUUGGCUUACACGCCCGCCCGAGAUUUUCUCAACCUUGAGUUUUUAAUUGGGCUAACGCCCGACUCGCUGGGC----ACUCGGUGGGCCAACGAGUG ..(((......)))..(((...(((((((((((.((...(((((((.....)))......(((((.......((((....)))))))))...))))----...))))))))))))).))) ( -46.21) >DroEre_CAF1 3115 112 + 1 ----UAAUAUGACCAACUUGGGUGUGGGCUUACACGCCCGGCCGAGAUUUUCUCAACCUUGAGUUUUUAAUUGGGCUAACGCCCGACUCGCUGGGC----AUUCGGUGGGCCAACGAGUG ----...........(((((......(((((((..(((((((.(((.....((((....)))).......((((((....))))))))))))))))----.....)))))))..))))). ( -45.10) >DroYak_CAF1 3227 116 + 1 ----UAAUAUGACCAACAUGGGUGUUGGCUUACACGCCCGCCCGAGAUUUUCUCAACCUUGAGUUUUUAAUUGGGCUAACGCCCGACUCGCUGGGCUCGCUUUUGGUGGGCCAACGAGUG ----..............(((((((((((......))).((((((......((((....)))).......)))))).))))))))(((((...(((((((.....)))))))..))))). ( -47.12) >consensus CAGGUAAUAUGACCAACAUGGGUGUUGGCUUACACGCCCGCCCGAGAUUUUCUCAACCUUGAGUUUUUAAUUGGGCUAACGCCCGACUCGCUGGGC____ACUCGGUGGGCCAACGAGUG ...........(((......)))((((((((((..((((...((((.....((((....)))).......((((((....))))))))))..)))).........))))))))))..... (-38.60 = -40.80 + 2.20)



| Location | 15,903,810 – 15,903,926 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.20 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -34.40 |
| Energy contribution | -34.44 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15903810 116 - 23771897 CACUCGUUGGCCCACCGAGU----GCCCAGCGAGUCGGGCGUUAGCCCAAUUAAAAACUCAAGGUUGAGAAAAUCUCGGCCGGGCGUGUAAGCCAACACCCAUGUUGGUCAUAUUACCUG .(((((((((..(((...))----).))))))))).((((....))))..............(((((((.....)))))))((..((((..(((((((....))))))).))))..)).. ( -44.10) >DroSec_CAF1 3225 116 - 1 CACUCGUUGGCCCACCGAGU----GCCCAGCGAGUCGGGCGUUAGCCCAAUUAAAAACUCAAGGUUGAGAAAAUCUCGGGCGGGCGUGUAAGCCAACACCCAUGUUGGUCAUAUUACCUG .(((((((((..(((...))----).))))))))).((((((..((((.......(((.....)))(((.....)))))))..))))....(((((((....))))))).......)).. ( -41.00) >DroSim_CAF1 5428 116 - 1 CACUCGUUGGCCCACCGAGU----GCCCAGCGAGUCGGGCGUUAGCCCAAUUAAAAACUCAAGGUUGAGAAAAUCUCGGGCGGGCGUGUAAGCCAACACCCAUGUUGGUCAUAUUACCUG .(((((((((..(((...))----).))))))))).((((((..((((.......(((.....)))(((.....)))))))..))))....(((((((....))))))).......)).. ( -41.00) >DroEre_CAF1 3115 112 - 1 CACUCGUUGGCCCACCGAAU----GCCCAGCGAGUCGGGCGUUAGCCCAAUUAAAAACUCAAGGUUGAGAAAAUCUCGGCCGGGCGUGUAAGCCCACACCCAAGUUGGUCAUAUUA---- .(((((((((.(........----).))))))))).((((....)))).......((((...(((((((.....)))))))(((.(((......))).))).))))..........---- ( -41.40) >DroYak_CAF1 3227 116 - 1 CACUCGUUGGCCCACCAAAAGCGAGCCCAGCGAGUCGGGCGUUAGCCCAAUUAAAAACUCAAGGUUGAGAAAAUCUCGGGCGGGCGUGUAAGCCAACACCCAUGUUGGUCAUAUUA---- .((.((((.(((((((....((.......))((((.((((....))))........))))..))).(((.....))))))).)))).))..(((((((....))))))).......---- ( -40.50) >consensus CACUCGUUGGCCCACCGAGU____GCCCAGCGAGUCGGGCGUUAGCCCAAUUAAAAACUCAAGGUUGAGAAAAUCUCGGGCGGGCGUGUAAGCCAACACCCAUGUUGGUCAUAUUACCUG (((...((((....))))......((((..((((..((((....)))).........((((....)))).....))))...)))))))...((((((......))))))........... (-34.40 = -34.44 + 0.04)


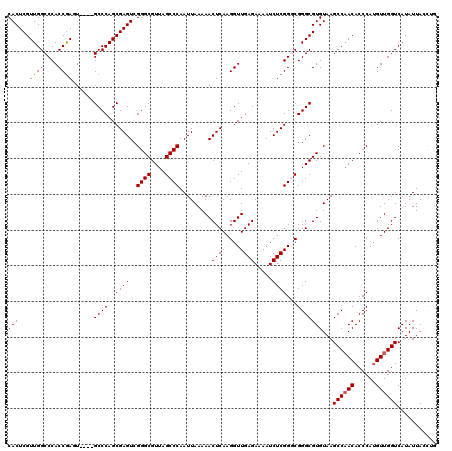
| Location | 15,903,926 – 15,904,042 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.62 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -30.98 |
| Energy contribution | -31.58 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15903926 116 + 23771897 ACUA-ACUUGCAGGAAAGGAAACACGGAGCGUGUGAGCCUCUCGCAUUUCACGUAAACACUUUGGCCAGAUACACUU---GUGGCCAAGAGCGACGGCUGAAAAUUUCUUUCUUUUCAUU ....-......(((((((((((((((...))))).((((..((((.......((....))((((((((.(......)---.)))))))).)))).)))).....))))))))))...... ( -35.10) >DroSec_CAF1 3341 119 + 1 ACUA-ACUGGCAGGAAAGGAAACACGGAGCGUGUGAGCCUCUCGCAUUUCACGUAAACACUUUGGCCAGAUACACUUGUUGUGGCCAAGAGCGACGGCUGAAAAUUUCUUUCUUUUCAUU ....-..(((.(((((((((((((((...))))).((((..((((.......((....))((((((((.(.((....))).)))))))).)))).)))).....)))))))))).))).. ( -37.10) >DroSim_CAF1 5544 120 + 1 ACUAAACUGGCAGGAAAGGAAACACGGAGCGUGUGAGCCUCUCGCAUUUCACGUAAACACUUUGGCCAGAUACACUUGUUGUGGCCAAGAGCGACGGCUGAAAAUUUCUUUCUUUUCAUU .......(((.(((((((((((((((...))))).((((..((((.......((....))((((((((.(.((....))).)))))))).)))).)))).....)))))))))).))).. ( -37.10) >DroEre_CAF1 3227 116 + 1 ACUA-ACUGGCAGGAAAGGAAACACGGAGCGUGUGAGCCUCUCGGAUUUCACGUAAACACUUUGGCCAGAUACACUU---GUGGCCAAGAGCGACGGCUGGAAAUUUCUUUCUUUUCAUU ....-..(((.((((((((((...(((..(((((((.((....))...)))).....(.(((((((((.(......)---.))))).)))).)))).)))....)))))))))).))).. ( -33.50) >DroYak_CAF1 3343 116 + 1 ACUA-ACUGGCAGGAAAGGAAACACGGAGCGUGUGAGCCUCUCGGAUUUCACGUAAACACUUUGGCCAGAUACACUU---GUGGCCAAGAGCGACGGCUGAAAAUUUCUUUCUUUUCAUA ....-..(((.(((((((((((((((...))))).((((..(((........((....))((((((((.(......)---.))))))))..))).)))).....)))))))))).))).. ( -33.50) >consensus ACUA_ACUGGCAGGAAAGGAAACACGGAGCGUGUGAGCCUCUCGCAUUUCACGUAAACACUUUGGCCAGAUACACUU___GUGGCCAAGAGCGACGGCUGAAAAUUUCUUUCUUUUCAUU .......(((.(((((((((((((((...))))).((((..((((.......((....))((((((((.............)))))))).)))).)))).....)))))))))).))).. (-30.98 = -31.58 + 0.60)

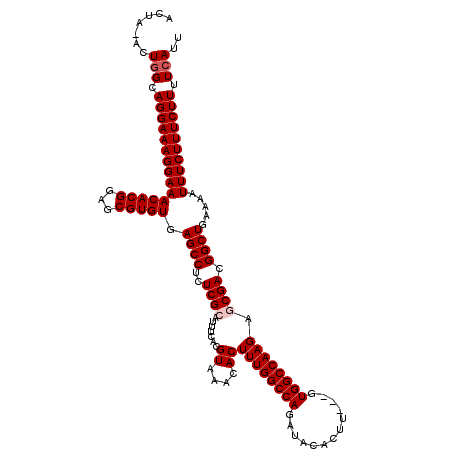
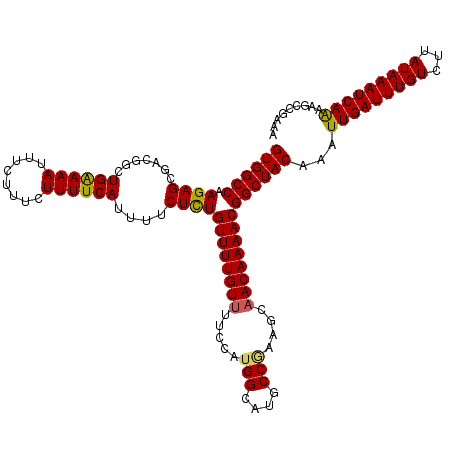
| Location | 15,904,002 – 15,904,122 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -28.52 |
| Energy contribution | -28.32 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15904002 120 + 23771897 GUGGCCAAGAGCGACGGCUGAAAAUUUCUUUCUUUUCAUUUUCUCUGUUUUGUUUUCCAUGGCAUGCCGAAGCAACAAAACGGCUACAAAUUGAUUUGUCUUAUAAAUCAAAAGCCGAAA ((((((.((((.((....((((((........)))))).)).))))((((((((.....(((....)))....))))))))))))))...(((((((((...)))))))))......... ( -29.70) >DroSec_CAF1 3420 120 + 1 GUGGCCAAGAGCGACGGCUGAAAAUUUCUUUCUUUUCAUUUUCUCUGUUUUGUAUUCCAUGGCAUGCCGAAGCAACAAAACGGCUACAAAUUGAUUUGUCUUAUAAAUCAGAAGCCGAAA ((((((.((((.((....((((((........)))))).)).))))(((((((.(((...((....)))))...)))))))))))))...(((((((((...)))))))))......... ( -28.50) >DroSim_CAF1 5624 120 + 1 GUGGCCAAGAGCGACGGCUGAAAAUUUCUUUCUUUUCAUUUUCUCUGUUUUGUUUUCCAUGGCAUGCCGAAGCAACAAAACGGCUACAAAUUGAUUUGUCUUAUAAAUCAAAAGCCGAAA ((((((.((((.((....((((((........)))))).)).))))((((((((.....(((....)))....))))))))))))))...(((((((((...)))))))))......... ( -29.70) >DroEre_CAF1 3303 120 + 1 GUGGCCAAGAGCGACGGCUGGAAAUUUCUUUCUUUUCAUUUUCUCUGUUUUGUUUUCCAUGGCAUGCCAAAACAACAAAACGGCUACAAAUUGAUUUGUCUUAUAAAUCAAAAGCCGAAA ((((((.((((.((.((..(((((....)))))..))..)).))))((((((((.....(((....)))....))))))))))))))...(((((((((...)))))))))......... ( -31.10) >DroYak_CAF1 3419 120 + 1 GUGGCCAAGAGCGACGGCUGAAAAUUUCUUUCUUUUCAUAUUCUUUGUUUUGUUUUCCACGGCAUGCCAAAACAACAAAACGGCUACAAAUUGAUUUGUCUUAUAAAUCAAAAGCCGAAA ((((((((((((....))((((((........))))))...)))).((((((((......((....)).....))))))))))))))...(((((((((...)))))))))......... ( -28.40) >consensus GUGGCCAAGAGCGACGGCUGAAAAUUUCUUUCUUUUCAUUUUCUCUGUUUUGUUUUCCAUGGCAUGCCGAAGCAACAAAACGGCUACAAAUUGAUUUGUCUUAUAAAUCAAAAGCCGAAA ((((((.((((.......((((((........))))))....))))((((((((.....(((....)))....))))))))))))))...(((((((((...)))))))))......... (-28.52 = -28.32 + -0.20)


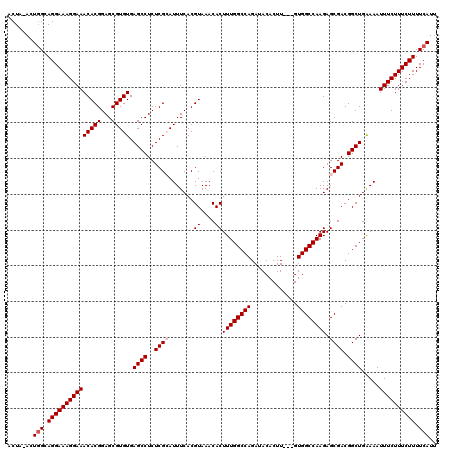
| Location | 15,904,002 – 15,904,122 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -26.38 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15904002 120 - 23771897 UUUCGGCUUUUGAUUUAUAAGACAAAUCAAUUUGUAGCCGUUUUGUUGCUUCGGCAUGCCAUGGAAAACAAAACAGAGAAAAUGAAAAGAAAGAAAUUUUCAGCCGUCGCUCUUGGCCAC ((((((((.(((((((.......))))))).....))))((((((((((....))...(....)..)))))))).))))...((((((........))))))((((.......))))... ( -29.90) >DroSec_CAF1 3420 120 - 1 UUUCGGCUUCUGAUUUAUAAGACAAAUCAAUUUGUAGCCGUUUUGUUGCUUCGGCAUGCCAUGGAAUACAAAACAGAGAAAAUGAAAAGAAAGAAAUUUUCAGCCGUCGCUCUUGGCCAC ((((((((..((((((.......))))))......))))(((((((..((..((....))..))...))))))).))))...((((((........))))))((((.......))))... ( -27.80) >DroSim_CAF1 5624 120 - 1 UUUCGGCUUUUGAUUUAUAAGACAAAUCAAUUUGUAGCCGUUUUGUUGCUUCGGCAUGCCAUGGAAAACAAAACAGAGAAAAUGAAAAGAAAGAAAUUUUCAGCCGUCGCUCUUGGCCAC ((((((((.(((((((.......))))))).....))))((((((((((....))...(....)..)))))))).))))...((((((........))))))((((.......))))... ( -29.90) >DroEre_CAF1 3303 120 - 1 UUUCGGCUUUUGAUUUAUAAGACAAAUCAAUUUGUAGCCGUUUUGUUGUUUUGGCAUGCCAUGGAAAACAAAACAGAGAAAAUGAAAAGAAAGAAAUUUCCAGCCGUCGCUCUUGGCCAC ((((((((.(((((((.......))))))).....))))((((((((....(((....))).....)))))))).)))).........((((....))))..((((.......))))... ( -30.20) >DroYak_CAF1 3419 120 - 1 UUUCGGCUUUUGAUUUAUAAGACAAAUCAAUUUGUAGCCGUUUUGUUGUUUUGGCAUGCCGUGGAAAACAAAACAAAGAAUAUGAAAAGAAAGAAAUUUUCAGCCGUCGCUCUUGGCCAC .(((((((.(((((((.......))))))).....))))((((((((....(((....))).....))))))))...)))..((((((........))))))((((.......))))... ( -29.10) >consensus UUUCGGCUUUUGAUUUAUAAGACAAAUCAAUUUGUAGCCGUUUUGUUGCUUCGGCAUGCCAUGGAAAACAAAACAGAGAAAAUGAAAAGAAAGAAAUUUUCAGCCGUCGCUCUUGGCCAC ((((((((.(((((((.......))))))).....))))(((((((..((..((....))..))...)))))))...)))).((((((........))))))((((.......))))... (-26.38 = -26.74 + 0.36)

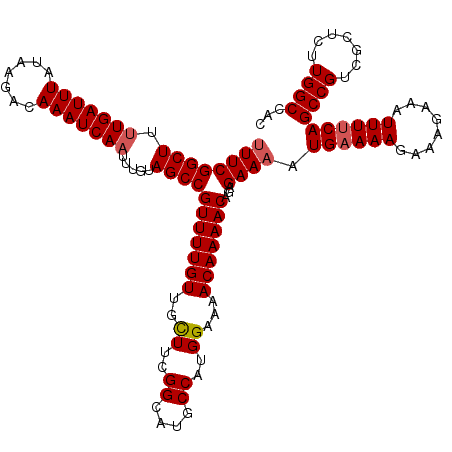
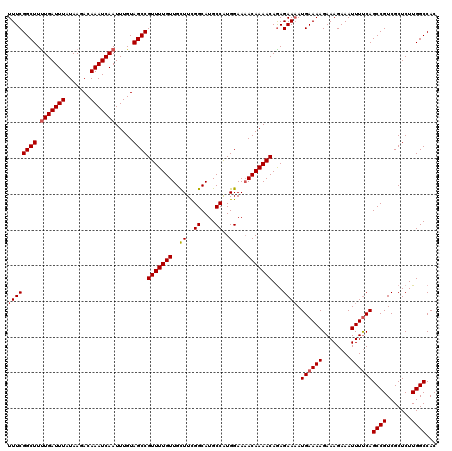
| Location | 15,904,042 – 15,904,162 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.16 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -27.24 |
| Energy contribution | -26.88 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15904042 120 - 23771897 ACUCCCCCGGCCGCUUCGGAUUCGAAAUGAUUUGAUUGCGUUUCGGCUUUUGAUUUAUAAGACAAAUCAAUUUGUAGCCGUUUUGUUGCUUCGGCAUGCCAUGGAAAACAAAACAGAGAA .(((....((((((.(((((((......)))))))..))).....((..(((((((.......)))))))...)).)))((((((((((....))...(....)..)))))))).))).. ( -30.00) >DroSec_CAF1 3460 120 - 1 ACUCCCCCGGCCGCUUCGAAUUCGAAAUGAUUUGAUUGCGUUUCGGCUUCUGAUUUAUAAGACAAAUCAAUUUGUAGCCGUUUUGUUGCUUCGGCAUGCCAUGGAAUACAAAACAGAGAA .(((....(((..(...(((..(((((((.........)))))))..)))((((((.......))))))....)..)))(((((((..((..((....))..))...))))))).))).. ( -31.60) >DroSim_CAF1 5664 120 - 1 ACUCCCCCGGCCGCAUCGAAUUCGAAAUGAUUUGAUUGCGUUUCGGCUUUUGAUUUAUAAGACAAAUCAAUUUGUAGCCGUUUUGUUGCUUCGGCAUGCCAUGGAAAACAAAACAGAGAA .(((....((((((((((((((......))))))).)))).....((..(((((((.......)))))))...)).)))((((((((((....))...(....)..)))))))).))).. ( -31.80) >DroEre_CAF1 3343 119 - 1 CCUCC-CCGGCCGCUUCGAAUUGGAAAUGAUUUGAUUGCGUUUCGGCUUUUGAUUUAUAAGACAAAUCAAUUUGUAGCCGUUUUGUUGUUUUGGCAUGCCAUGGAAAACAAAACAGAGAA .(((.-..((((((.(((((((......)))))))..))).....((..(((((((.......)))))))...)).)))((((((((....(((....))).....)))))))).))).. ( -31.80) >DroYak_CAF1 3459 119 - 1 CCUCC-CCGGCCGCUUCGAAUUGGAAAUGGUUUGAUUGCGUUUCGGCUUUUGAUUUAUAAGACAAAUCAAUUUGUAGCCGUUUUGUUGUUUUGGCAUGCCGUGGAAAACAAAACAAAGAA ..(((-.(((((((.(((((((......)))))))..)))...(((((.(((((((.......))))))).....)))))...((((.....)))).)))).)))............... ( -29.50) >consensus ACUCCCCCGGCCGCUUCGAAUUCGAAAUGAUUUGAUUGCGUUUCGGCUUUUGAUUUAUAAGACAAAUCAAUUUGUAGCCGUUUUGUUGCUUCGGCAUGCCAUGGAAAACAAAACAGAGAA ..(((...((((((.(((((((......)))))))..)))...(((((.(((((((.......))))))).....)))))......(((....))).)))..)))............... (-27.24 = -26.88 + -0.36)

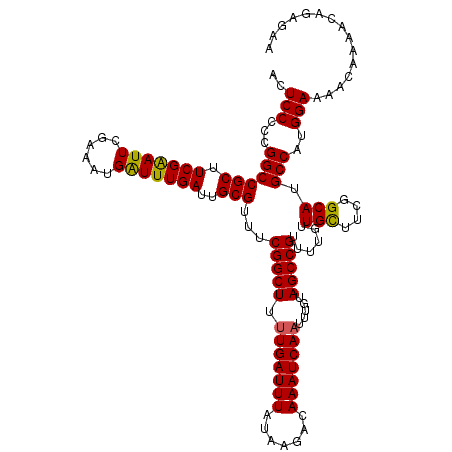

| Location | 15,904,082 – 15,904,202 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.16 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -31.66 |
| Energy contribution | -32.30 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15904082 120 + 23771897 CGGCUACAAAUUGAUUUGUCUUAUAAAUCAAAAGCCGAAACGCAAUCAAAUCAUUUCGAAUCCGAAGCGGCCGGGGGAGUAGUGGGAAAAGUUUCGCUUUCUGCAGCUGCUUUUAGUUUU (((((.....(((((((((...))))))))).)))))....................((((..((((((((..(.((((.((((..(....)..)))))))).).))))))))..)))). ( -33.40) >DroSec_CAF1 3500 120 + 1 CGGCUACAAAUUGAUUUGUCUUAUAAAUCAGAAGCCGAAACGCAAUCAAAUCAUUUCGAAUUCGAAGCGGCCGGGGGAGUAGUGGGAAAAGUUUCGCUUUCUGCAGCUGCUUUUAGUUUU (((((.....(((((((((...))))))))).)))))....................(((((.((((((((..(.((((.((((..(....)..)))))))).).)))))))).))))). ( -34.30) >DroSim_CAF1 5704 120 + 1 CGGCUACAAAUUGAUUUGUCUUAUAAAUCAAAAGCCGAAACGCAAUCAAAUCAUUUCGAAUUCGAUGCGGCCGGGGGAGUAGUGGGAAAAGUUUCGCUUUCUGCAGCUGCUUUUAGUUUU (((((.....(((((((((...))))))))).)))))((((..............(((....))).(((((..(.((((.((((..(....)..)))))))).).))))).....)))). ( -32.00) >DroEre_CAF1 3383 119 + 1 CGGCUACAAAUUGAUUUGUCUUAUAAAUCAAAAGCCGAAACGCAAUCAAAUCAUUUCCAAUUCGAAGCGGCCGG-GGAGGAGUGGGAAAAGUUUCGCUUUCUGCAGCUGCUUUUAGUUUU (((((.....(((((((((...))))))))).))))).....................((((.((((((((.(.-((((.((((..(....)..)))))))).).)))))))).)))).. ( -34.50) >DroYak_CAF1 3499 119 + 1 CGGCUACAAAUUGAUUUGUCUUAUAAAUCAAAAGCCGAAACGCAAUCAAACCAUUUCCAAUUCGAAGCGGCCGG-GGAGGAGUGGGAAAAGUUUCGCUUUCUGCAGCUGCUUUUAGUUUU (((((.....(((((((((...))))))))).))))).....................((((.((((((((.(.-((((.((((..(....)..)))))))).).)))))))).)))).. ( -34.50) >consensus CGGCUACAAAUUGAUUUGUCUUAUAAAUCAAAAGCCGAAACGCAAUCAAAUCAUUUCGAAUUCGAAGCGGCCGGGGGAGUAGUGGGAAAAGUUUCGCUUUCUGCAGCUGCUUUUAGUUUU (((((.....(((((((((...))))))))).)))))....................(((((.((((((((..(.((((.((((..(....)..)))))))).).)))))))).))))). (-31.66 = -32.30 + 0.64)

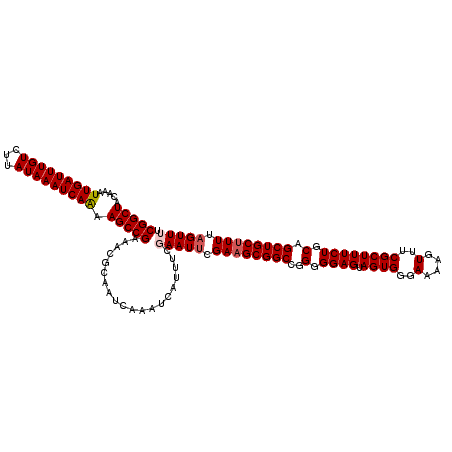
| Location | 15,904,162 – 15,904,275 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.40 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -21.58 |
| Energy contribution | -21.58 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15904162 113 - 23771897 AAAAUCACGGCUAAUUAUUGUUGGCAUGA----ACGGGAACACCGAACAUCGCGUAC---ACACAGGGAAAACGUGCUGAAAAACUAAAAGCAGCUGCAGAAAGCGAAACUUUUCCCACU ......(((((((((....))))))....----.(((.....))).......)))..---.....(((((((.(((((...........))).(((......)))...)))))))))... ( -27.20) >DroSec_CAF1 3580 109 - 1 AAAAUCACGGCUAAUUAUUGUUGGCAUGA----ACGGCAACACC-------GCGUACACCACACAGGGAAAACGUGCUGAAAAACUAAAAGCAGCUGCAGAAAGCGAAACUUUUCCCACU ......(((((((((....))))))....----.(((.....))-------))))..........(((((((.(((((...........))).(((......)))...)))))))))... ( -25.20) >DroSim_CAF1 5784 97 - 1 AAAAUCACGGCUAAUUAUUGUUGGCAUGA----ACGGGAAC-------------------ACACAGGGAAAACGUGCUGAAAAACUAAAAGCAGCUGCAGAAAGCGAAACUUUUCCCACU ....(((..((((((....)))))).)))----..(....)-------------------.....(((((((.(((((...........))).(((......)))...)))))))))... ( -22.40) >DroEre_CAF1 3462 117 - 1 AAAAUCACGGCUAAUUAUGGUUGGCAUGAACGAACGGGAACACCGACCAUUGCGUAC---AUACAGGGAAAACGUGCUGAAAAACUAAAAGCAGCUGCAGAAAGCGAAACUUUUCCCACU ......(((((((((....)))))).........(((.....))).......)))..---.....(((((((.(((((...........))).(((......)))...)))))))))... ( -27.00) >DroYak_CAF1 3578 113 - 1 AAAAUCACGGCUAAUUAUUGUUGGCAUGA----ACGGGAACACCGAACAUCGCGUAC---ACACAGGGAAAACGUGCUGAAAAACUAAAAGCAGCUGCAGAAAGCGAAACUUUUCCCACU ......(((((((((....))))))....----.(((.....))).......)))..---.....(((((((.(((((...........))).(((......)))...)))))))))... ( -27.20) >consensus AAAAUCACGGCUAAUUAUUGUUGGCAUGA____ACGGGAACACCGA_CAU_GCGUAC___ACACAGGGAAAACGUGCUGAAAAACUAAAAGCAGCUGCAGAAAGCGAAACUUUUCCCACU ....(((..((((((....)))))).)))......(....)........................(((((((.(((((...........))).(((......)))...)))))))))... (-21.58 = -21.58 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:59 2006