
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,894,216 – 15,894,331 |
| Length | 115 |
| Max. P | 0.999969 |

| Location | 15,894,216 – 15,894,331 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -28.50 |
| Energy contribution | -28.34 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
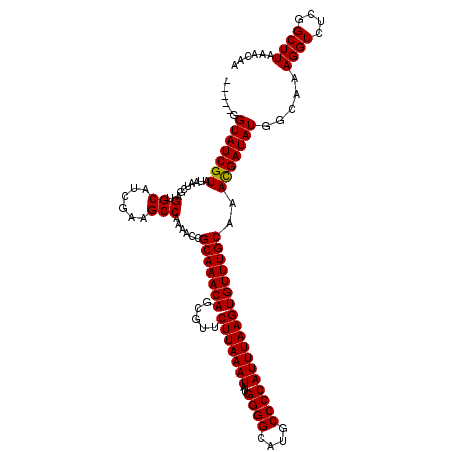
>3L_DroMel_CAF1 15894216 115 + 23771897 -----GGUAUCGUAUAAUCGAGUGCAUCGAAGCCAAAACCGCAAACAGCGUUCUUAAAUAUCGGGGCAUGCCCCAUUUAAGUGUUUGCAAACGAUAUGGCAAAGGUCUCAGCUUAAACAA -----.(((((((....((((.....))))..........(((((((.....(((((((...((((....))))))))))))))))))..)))))))(((....)))............. ( -30.90) >DroSec_CAF1 2732 115 + 1 -----GGUAUCGUAUAAUCGAGUGCAUCGAAGCCAAAACCGCAAACAGCGUUCUUAAAUAUCGGGGCAUGCCCCAUUUAAGUGUUUGCAAACGAUAUGGCAAAGGUCUCGGCUUAAACAA -----.(((((((....((((.....))))..........(((((((.....(((((((...((((....))))))))))))))))))..))))))).....((((....))))...... ( -31.70) >DroSim_CAF1 4941 115 + 1 -----GGUAUCGUAUAAUCGAGUGCAUCGAAGCCAAAACCGCAAACAGCGUUCUUAAAUAUCGGGGCAUGCCCCAUUUAAGUGUUUGCAAACGAUAUGGCAAAGGUCUCAGCUUAAACAA -----.(((((((....((((.....))))..........(((((((.....(((((((...((((....))))))))))))))))))..)))))))(((....)))............. ( -30.90) >DroEre_CAF1 2687 120 + 1 GGUAGGGUAUCGUAUUAUCGAGUGCAGCCAAGCCAAAACCGCAAACAACGUUCUUAAAUAUCGGGGCAUGCCCCAUUUAAGUGUUUGCAAACGAUAUGGCAAAGGUCUCGGCUUAACCAA (((..(((...(((((....))))).)))(((((......(((((((.....(((((((...((((....)))))))))))))))))).........(((....)))..))))).))).. ( -35.60) >DroYak_CAF1 2774 115 + 1 -----GGUAUCGUAUUAUCGAGAGCAGCGAAGCCAAAACCGCAAACAGCGUUCUUAAAUAUCGGGGCAUGCCCCAUUUAAGUGUUUGCAAAUGAUAUGGCAAAGGUCUCGGCUUAACCAA -----(((.(((......)))((((..(((..((......(((((((.....(((((((...((((....))))))))))))))))))...((......))..))..))))))).))).. ( -33.60) >consensus _____GGUAUCGUAUAAUCGAGUGCAUCGAAGCCAAAACCGCAAACAGCGUUCUUAAAUAUCGGGGCAUGCCCCAUUUAAGUGUUUGCAAACGAUAUGGCAAAGGUCUCGGCUUAAACAA ......(((((((........(.((......)))......(((((((.....(((((((...((((....))))))))))))))))))..))))))).....((((....))))...... (-28.50 = -28.34 + -0.16)


| Location | 15,894,216 – 15,894,331 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -36.06 |
| Consensus MFE | -35.30 |
| Energy contribution | -34.42 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.02 |
| SVM RNA-class probability | 0.999969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15894216 115 - 23771897 UUGUUUAAGCUGAGACCUUUGCCAUAUCGUUUGCAAACACUUAAAUGGGGCAUGCCCCGAUAUUUAAGAACGCUGUUUGCGGUUUUGGCUUCGAUGCACUCGAUUAUACGAUACC----- ((((.(((((..(((((...((......))..((((((((((((((((((....))))...))))))).....))))))))))))..)).((((.....))))))).))))....----- ( -33.30) >DroSec_CAF1 2732 115 - 1 UUGUUUAAGCCGAGACCUUUGCCAUAUCGUUUGCAAACACUUAAAUGGGGCAUGCCCCGAUAUUUAAGAACGCUGUUUGCGGUUUUGGCUUCGAUGCACUCGAUUAUACGAUACC----- ((((.((((((((((((...((......))..((((((((((((((((((....))))...))))))).....)))))))))))))))).((((.....))))))).))))....----- ( -35.80) >DroSim_CAF1 4941 115 - 1 UUGUUUAAGCUGAGACCUUUGCCAUAUCGUUUGCAAACACUUAAAUGGGGCAUGCCCCGAUAUUUAAGAACGCUGUUUGCGGUUUUGGCUUCGAUGCACUCGAUUAUACGAUACC----- ((((.(((((..(((((...((......))..((((((((((((((((((....))))...))))))).....))))))))))))..)).((((.....))))))).))))....----- ( -33.30) >DroEre_CAF1 2687 120 - 1 UUGGUUAAGCCGAGACCUUUGCCAUAUCGUUUGCAAACACUUAAAUGGGGCAUGCCCCGAUAUUUAAGAACGUUGUUUGCGGUUUUGGCUUGGCUGCACUCGAUAAUACGAUACCCUACC ..(((((((((((((((...((......))..((((((((((((((((((....))))...))))))).....))))))))))))))))))))))....(((......)))......... ( -40.70) >DroYak_CAF1 2774 115 - 1 UUGGUUAAGCCGAGACCUUUGCCAUAUCAUUUGCAAACACUUAAAUGGGGCAUGCCCCGAUAUUUAAGAACGCUGUUUGCGGUUUUGGCUUCGCUGCUCUCGAUAAUACGAUACC----- ..(((.(((((((((((..((......))...((((((((((((((((((....))))...))))))).....)))))))))))))))))).)))....(((......)))....----- ( -37.20) >consensus UUGUUUAAGCCGAGACCUUUGCCAUAUCGUUUGCAAACACUUAAAUGGGGCAUGCCCCGAUAUUUAAGAACGCUGUUUGCGGUUUUGGCUUCGAUGCACUCGAUUAUACGAUACC_____ ..((..(((((((((((..((......))...((((((((((((((((((....))))...))))))).....))))))))))))))))))..))....(((......)))......... (-35.30 = -34.42 + -0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:49 2006