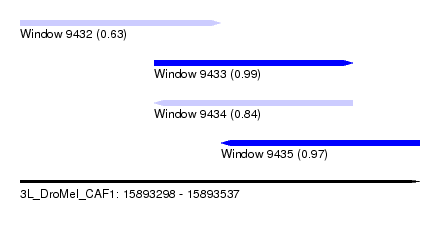
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,893,298 – 15,893,537 |
| Length | 239 |
| Max. P | 0.987339 |

| Location | 15,893,298 – 15,893,418 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -26.02 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15893298 120 + 23771897 UACUCGGGCGUUCCACACAAUGUCCAAGUGCGAUCCCUCAGCUUUUUGGCAAAUCCAAAAUCGGUUAUCUUCAAAUGACCAUCCCGAUUGAUCAGCAGAUUCUCUGGCUUCAGAUCGCGA .(((.(((((((......))))))).)))((((((....(((.((((((.....))))))..((((((......))))))...............(((.....))))))...)))))).. ( -32.20) >DroSec_CAF1 1818 120 + 1 UACUCGGGCGUUCCGCACAAUGUCCAAGUGCGAUCCCUGAGCUUUUUGGCAAAGCCAAAAUCGGUUAUCUUCAAAUGGCCAUCGCGAUUGAUCAGCAGAUUCUCUGGCUUCAGAUCGCGA ..((((((.(...(((((.........)))))..)))))))..(((((((...)))))))..((((((......)))))).((((((((....(((((.....)).)))...)))))))) ( -41.20) >DroSim_CAF1 4031 120 + 1 UACUCGGGCGUUCCGCACAAUGUCCAAGUGCGAUCCCUCAGCUUUUUGGCAAAGCCAAAAUCGGUUAUCUUCAAAUGGCCAUCGCGAUUGAUCAGCAGAUUCUCUGGCUUCAGAUCGCGA .(((.(((((((......))))))).)))((((((.....(((....))).((((((.((((((((((......))))))...((.........)).))))...))))))..)))))).. ( -37.40) >DroEre_CAF1 1794 120 + 1 UACUCGGGCGUUCCGCACAAUGUCCAAGUGCGAUCCCUUAGUUUUUUGGCAAAGCCAAAAUCUGUUAUUUUUAAAUGGCCAUCCCGAUUGAUCAGCAGAUUCUCUGGCUUUAAGUCGCGG .(((.(((((((......))))))).)))(((((......(((....)))(((((((.((((((((....((((.(((.....))).))))..))))))))...)))))))..))))).. ( -34.40) >DroYak_CAF1 1858 120 + 1 UACUCAGGUGUUCCGCACAAAGUCCAAGUGCGAUCCCUCAGCUUUUUGGCAAAGCCGAAAUCUGUUAUUUUUAAAUGGCCAUCCCUAUUGAUCAGCAGAUUCUCUGGCUUUAAGUCACGA ..((.(((.(...(((((.........)))))..)))).)).....((((((((((((((((((((.......(((((......)))))....)))))))).)).))))))..))))... ( -29.20) >consensus UACUCGGGCGUUCCGCACAAUGUCCAAGUGCGAUCCCUCAGCUUUUUGGCAAAGCCAAAAUCGGUUAUCUUCAAAUGGCCAUCCCGAUUGAUCAGCAGAUUCUCUGGCUUCAGAUCGCGA .(((.(((((((......))))))).)))((((((....(((.(((((((...)))))))..((((((......))))))...............(((.....))))))...)))))).. (-26.02 = -26.74 + 0.72)


| Location | 15,893,378 – 15,893,497 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -29.80 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15893378 119 + 23771897 AUCCCGAUUGAUCAGCAGAUUCUCUGGCUUCAGAUCGCGAUAAACAAUUUGCAGUGAGUGCAGGUAUUCUAG-GGCACUGACGAUCUCAGCCGCAUAGAAGUUGGCUAAAAUCGAAAUCA ....(((((....(((...((((.(((((..(((((((((........)))).((.(((((.((....))..-.))))).))))))).)))))...))))....)))..)))))...... ( -36.30) >DroSec_CAF1 1898 120 + 1 AUCGCGAUUGAUCAGCAGAUUCUCUGGCUUCAGAUCGCGAUAAACAAUUUGCAGUGAGUGCAGGUAUUCCAGGGGCACUCACGAUCUCAGCCGCAUAGAAAUUGGCUAAAAUCGAAAUCA ....(((((....(((...((((.(((((..(((((((((........)))).((((((((.((....))....))))))))))))).)))))...))))....)))..)))))...... ( -40.50) >DroSim_CAF1 4111 119 + 1 AUCGCGAUUGAUCAGCAGAUUCUCUGGCUUCAGAUCGCGAUAAACAAUUUGCAGUGAGUGCAGGUAUUCCAG-CGCACUCACGAUCUCAGCCGCAUAGAAAUUGGCUAAAAUCGAAAUCA ....(((((....(((...((((.(((((..(((((((((........)))).((((((((.((....))..-.))))))))))))).)))))...))))....)))..)))))...... ( -40.00) >DroEre_CAF1 1874 118 + 1 AUCCCGAUUGAUCAGCAGAUUCUCUGGCUUUAAGUCGCGGUAAACAAUUUGCAGUGAGUGCAGGUAUUCCAG-GGCACUGACGAUCUCAGCCGCAUAGAAGUUGGCUGAAA-CAAAAUGA .........((((.(((((((((.((((.....)))).)).....))))))).((.(((((.((....))..-.))))).))))))((((((((......).)))))))..-........ ( -35.00) >DroYak_CAF1 1938 119 + 1 AUCCCUAUUGAUCAGCAGAUUCUCUGGCUUUAAGUCACGAUAAACAAUUUGCAGUGAGUGCAGGUAUUCCAG-GGCACUGACGAUCUCAGCCGCAUAGAAGUUGGCUAAAAUCAAAAUCA .........((((.(((((((.((((((.....)))).)).....))))))).((.(((((.((....))..-.))))).))))))..((((((......).)))))............. ( -30.70) >consensus AUCCCGAUUGAUCAGCAGAUUCUCUGGCUUCAGAUCGCGAUAAACAAUUUGCAGUGAGUGCAGGUAUUCCAG_GGCACUGACGAUCUCAGCCGCAUAGAAGUUGGCUAAAAUCGAAAUCA .......(((((.(((...((((.(((((..(((((((((........)))).((.(((((.((....))....))))).))))))).)))))...))))....)))...)))))..... (-29.80 = -29.80 + 0.00)


| Location | 15,893,378 – 15,893,497 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -27.64 |
| Energy contribution | -28.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15893378 119 - 23771897 UGAUUUCGAUUUUAGCCAACUUCUAUGCGGCUGAGAUCGUCAGUGCC-CUAGAAUACCUGCACUCACUGCAAAUUGUUUAUCGCGAUCUGAAGCCAGAGAAUCUGCUGAUCAAUCGGGAU ..((..((((((((((....((((....((((.(((((((.(((((.-..((.....))))))).)).((............)))))))..))))..))))...)))))..)))))..)) ( -32.60) >DroSec_CAF1 1898 120 - 1 UGAUUUCGAUUUUAGCCAAUUUCUAUGCGGCUGAGAUCGUGAGUGCCCCUGGAAUACCUGCACUCACUGCAAAUUGUUUAUCGCGAUCUGAAGCCAGAGAAUCUGCUGAUCAAUCGCGAU .((((((((((((((((...........))))))))))((((((((....((....)).)))))))).).)))))....((((((((.((((((.((.....)))))..))))))))))) ( -43.00) >DroSim_CAF1 4111 119 - 1 UGAUUUCGAUUUUAGCCAAUUUCUAUGCGGCUGAGAUCGUGAGUGCG-CUGGAAUACCUGCACUCACUGCAAAUUGUUUAUCGCGAUCUGAAGCCAGAGAAUCUGCUGAUCAAUCGCGAU .((((((((((((((((...........))))))))))(((((((((-..((....))))))))))).).)))))....((((((((.((((((.((.....)))))..))))))))))) ( -43.70) >DroEre_CAF1 1874 118 - 1 UCAUUUUG-UUUCAGCCAACUUCUAUGCGGCUGAGAUCGUCAGUGCC-CUGGAAUACCUGCACUCACUGCAAAUUGUUUACCGCGACUUAAAGCCAGAGAAUCUGCUGAUCAAUCGGGAU .((.((((-((((((((...........)))))))...((.(((((.-..((....)).))))).)).))))).))....((.(((.....(((.((.....)))))......))))).. ( -29.30) >DroYak_CAF1 1938 119 - 1 UGAUUUUGAUUUUAGCCAACUUCUAUGCGGCUGAGAUCGUCAGUGCC-CUGGAAUACCUGCACUCACUGCAAAUUGUUUAUCGUGACUUAAAGCCAGAGAAUCUGCUGAUCAAUAGGGAU ((((...((((((((((...........))))))))))))))...((-((((((((..((((.....))))...)))))....(((.....(((.((.....)))))..))).))))).. ( -28.00) >consensus UGAUUUCGAUUUUAGCCAACUUCUAUGCGGCUGAGAUCGUCAGUGCC_CUGGAAUACCUGCACUCACUGCAAAUUGUUUAUCGCGAUCUGAAGCCAGAGAAUCUGCUGAUCAAUCGGGAU ..((((((((((((((....((((....((((.(((((((.(((((....((....)).))))).)).((............)))))))..))))..))))...)))))..))))))))) (-27.64 = -28.24 + 0.60)

| Location | 15,893,418 – 15,893,537 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.18 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15893418 119 - 23771897 AACUAUUCUAUUAAAUCUUUUAAAUCAGAAAUCACAAAAUUGAUUUCGAUUUUAGCCAACUUCUAUGCGGCUGAGAUCGUCAGUGCC-CUAGAAUACCUGCACUCACUGCAAAUUGUUUA ...(((((((.................(((((((......)))))))((((((((((...........)))))))))).........-.)))))))..((((.....))))......... ( -27.30) >DroSec_CAF1 1938 120 - 1 AACUAUUCUAUUAAAUCUUCUAAAUAAAAAAUCACAAAACUGAUUUCGAUUUUAGCCAAUUUCUAUGCGGCUGAGAUCGUGAGUGCCCCUGGAAUACCUGCACUCACUGCAAAUUGUUUA ....................(((((((.((((((......)))))).((((((((((...........))))))))))((((((((....((....)).))))))))......))))))) ( -31.30) >DroSim_CAF1 4151 119 - 1 AACUAUUAAAUUAAAUCUUUUAAAUCAAAAAUCACUAAACUGAUUUCGAUUUUAGCCAAUUUCUAUGCGGCUGAGAUCGUGAGUGCG-CUGGAAUACCUGCACUCACUGCAAAUUGUUUA .....(((((........)))))............(((((.((((((((((((((((...........))))))))))(((((((((-..((....))))))))))).).)))))))))) ( -30.80) >DroEre_CAF1 1914 117 - 1 CACCAUUCAA-UAUAUCUUUUAUGUAUGUAUCAAUAGAAAUCAUUUUG-UUUCAGCCAACUUCUAUGCGGCUGAGAUCGUCAGUGCC-CUGGAAUACCUGCACUCACUGCAAAUUGUUUA .........(-((((.....))))).((((.................(-((((((((...........))))))))).((.(((((.-..((....)).))))).))))))......... ( -22.50) >DroYak_CAF1 1978 119 - 1 GGCCAUUCUAUUAAAUCGUUUAUGUAUGUAUCAAUAAUAAUGAUUUUGAUUUUAGCCAACUUCUAUGCGGCUGAGAUCGUCAGUGCC-CUGGAAUACCUGCACUCACUGCAAAUUGUUUA .((.........((((((((....(((......)))..)))))))).((((((((((...........))))))))))((.(((((.-..((....)).))))).)).)).......... ( -27.20) >consensus AACUAUUCUAUUAAAUCUUUUAAAUAAGAAAUCACAAAAAUGAUUUCGAUUUUAGCCAACUUCUAUGCGGCUGAGAUCGUCAGUGCC_CUGGAAUACCUGCACUCACUGCAAAUUGUUUA ...............................................((((((((((...........))))))))))((.(((((....((....)).))))).))............. (-19.94 = -20.18 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:47 2006