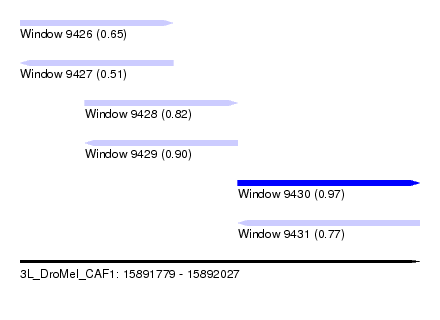
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,891,779 – 15,892,027 |
| Length | 248 |
| Max. P | 0.973747 |

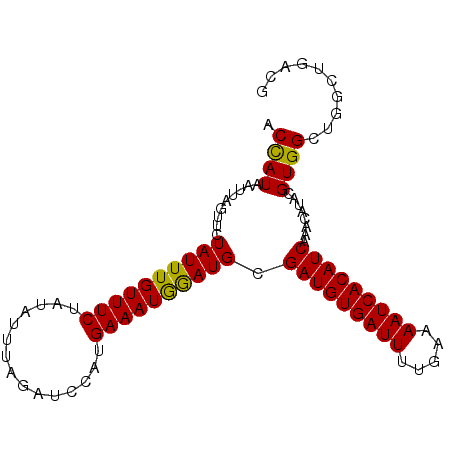
| Location | 15,891,779 – 15,891,874 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -21.17 |
| Consensus MFE | -19.39 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15891779 95 + 23771897 ACCAUAAUUAGUUCUAUUUGUUUCUAUAUUUAGAUCCAUGAAAUAGAUGUGAUGUGAUUUUGAAAAUCACAUCAAACAUACGUGGCUGGCAAACG .((((.......((((((((..((((....))))..))...))))))..((((((((((.....)))))))))).......)))).......... ( -22.70) >DroSec_CAF1 297 95 + 1 ACUAUAAUAAGUUCUAUUUGUUUCUAAAUUUAGAUCCAUGAAAUGGAUGCGAUGUGAUUUUGAAAAUCACAUCAAACAUACGUGGCUGGCUGACG .........((((((((.(((((..........((((((...))))))..(((((((((.....))))))))))))))...))))..)))).... ( -20.20) >DroSim_CAF1 2493 95 + 1 ACCAUAAUUAGUUCUAUUUGUUUCUAUAUUUAGAUCCAUGAAAUGAAUGCGAUGUGAUUUUGAAAAUCACAUCAAACAUACGUGGCUGGCUGACG .((((.....((((....((..((((....))))..))......))))..(((((((((.....)))))))))........)))).......... ( -20.60) >consensus ACCAUAAUUAGUUCUAUUUGUUUCUAUAUUUAGAUCCAUGAAAUGGAUGCGAUGUGAUUUUGAAAAUCACAUCAAACAUACGUGGCUGGCUGACG .((((.........((((((((((...............)))))))))).(((((((((.....)))))))))........)))).......... (-19.39 = -18.73 + -0.66)


| Location | 15,891,779 – 15,891,874 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -19.33 |
| Consensus MFE | -16.57 |
| Energy contribution | -16.47 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15891779 95 - 23771897 CGUUUGCCAGCCACGUAUGUUUGAUGUGAUUUUCAAAAUCACAUCACAUCUAUUUCAUGGAUCUAAAUAUAGAAACAAAUAGAACUAAUUAUGGU .....((((............((((((((((.....))))))))))..((((((...((..((((....))))..))))))))........)))) ( -21.60) >DroSec_CAF1 297 95 - 1 CGUCAGCCAGCCACGUAUGUUUGAUGUGAUUUUCAAAAUCACAUCGCAUCCAUUUCAUGGAUCUAAAUUUAGAAACAAAUAGAACUUAUUAUAGU .................((((((((((((((.....)))))))))(.((((((...))))))).........))))).................. ( -17.30) >DroSim_CAF1 2493 95 - 1 CGUCAGCCAGCCACGUAUGUUUGAUGUGAUUUUCAAAAUCACAUCGCAUUCAUUUCAUGGAUCUAAAUAUAGAAACAAAUAGAACUAAUUAUGGU .....((((.....(.((((..(((((((((.....))))))))))))).)......((..((((....))))..))..............)))) ( -19.10) >consensus CGUCAGCCAGCCACGUAUGUUUGAUGUGAUUUUCAAAAUCACAUCGCAUCCAUUUCAUGGAUCUAAAUAUAGAAACAAAUAGAACUAAUUAUGGU .....((((.....(.((((..(((((((((.....))))))))))))).)......((..((((....))))..))..............)))) (-16.57 = -16.47 + -0.11)

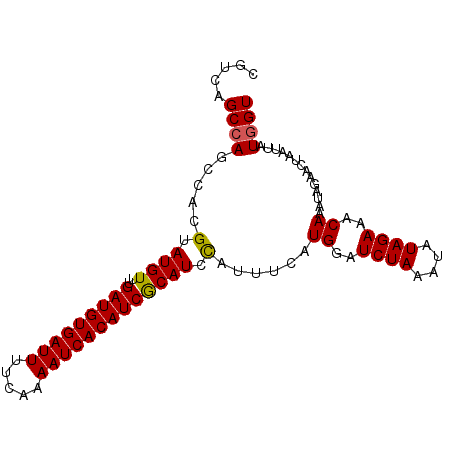

| Location | 15,891,819 – 15,891,914 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -13.97 |
| Energy contribution | -14.84 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.819255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15891819 95 + 23771897 AAAUAGA-UGUG--------------------AUGUGAUUUUGAAAAUCACAUCAAACAUACGUGGCUGGCAAACGGAUAAAAGUUGCGAAAUGUGCGCAAAAAUUCAACACGUUU ......(-((((--------------------((((((((.....)))))))))..))))(((((.(((.....))).......(((((.......)))))........))))).. ( -23.90) >DroSec_CAF1 337 94 + 1 AAAUGGA-UGCG--------------------AUGUGAUUUUGAAAAUCACAUCAAACAUACGUGGCUGGCUGACGGAUAAAAGUUGCGAAAUGUGCGCAA-AAUUCAGCACGUUU .......-...(--------------------((((((((.....))))))))).......((..(((..............)))..))((((((((....-......)))))))) ( -25.94) >DroSim_CAF1 2533 94 + 1 AAAUGAA-UGCG--------------------AUGUGAUUUUGAAAAUCACAUCAAACAUACGUGGCUGGCUGACGGAUAAAAGUUGCGAAAUGUGCGCAA-AAUUCAGCACGUUU .......-...(--------------------((((((((.....))))))))).......((..(((..............)))..))((((((((....-......)))))))) ( -25.94) >DroYak_CAF1 317 110 + 1 AAAUGGAAUUCCAAGCUAUAGCAUUUUCAUUUUUGUGA-UUUGAAAAUCACAUCAAACAUA----CGUAGCCGACGGGUAAAAUUUGCGAAAUGUGCGCAA-AAUUCAGCACGUUU ...(((....))).......(((..........(((((-((....))))))).........----....(((....)))......))).((((((((....-......)))))))) ( -21.60) >consensus AAAUGGA_UGCG____________________AUGUGAUUUUGAAAAUCACAUCAAACAUACGUGGCUGGCUGACGGAUAAAAGUUGCGAAAUGUGCGCAA_AAUUCAGCACGUUU .................................(((((((.....))))))).........((..(((..............)))..))((((((((...........)))))))) (-13.97 = -14.84 + 0.87)


| Location | 15,891,819 – 15,891,914 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -13.19 |
| Energy contribution | -13.19 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15891819 95 - 23771897 AAACGUGUUGAAUUUUUGCGCACAUUUCGCAACUUUUAUCCGUUUGCCAGCCACGUAUGUUUGAUGUGAUUUUCAAAAUCACAU--------------------CACA-UCUAUUU (((((.(.((((...(((((.......)))))..)))).)))))).........(((.((.((((((((((.....))))))))--------------------)).)-).))).. ( -20.40) >DroSec_CAF1 337 94 - 1 AAACGUGCUGAAUU-UUGCGCACAUUUCGCAACUUUUAUCCGUCAGCCAGCCACGUAUGUUUGAUGUGAUUUUCAAAAUCACAU--------------------CGCA-UCCAUUU ..((((((((....-(((((.......))))).........(....)))).)))))((((..(((((((((.....))))))))--------------------))))-)...... ( -22.50) >DroSim_CAF1 2533 94 - 1 AAACGUGCUGAAUU-UUGCGCACAUUUCGCAACUUUUAUCCGUCAGCCAGCCACGUAUGUUUGAUGUGAUUUUCAAAAUCACAU--------------------CGCA-UUCAUUU ..((((((((....-(((((.......))))).........(....)))).)))))((((..(((((((((.....))))))))--------------------))))-)...... ( -22.50) >DroYak_CAF1 317 110 - 1 AAACGUGCUGAAUU-UUGCGCACAUUUCGCAAAUUUUACCCGUCGGCUACG----UAUGUUUGAUGUGAUUUUCAAA-UCACAAAAAUGAAAAUGCUAUAGCUUGGAAUUCCAUUU ..(((((((((..(-(((((.......)))))).........))))).)))----)..((((..(((((((....))-))))).))))......((....)).(((....)))... ( -23.50) >consensus AAACGUGCUGAAUU_UUGCGCACAUUUCGCAACUUUUAUCCGUCAGCCAGCCACGUAUGUUUGAUGUGAUUUUCAAAAUCACAU____________________CGCA_UCCAUUU (((((((((((....(((((.......)))))..........))))).........))))))..(((((((.....)))))))................................. (-13.19 = -13.19 + 0.00)



| Location | 15,891,914 – 15,892,027 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.21 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -28.28 |
| Energy contribution | -28.04 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15891914 113 + 23771897 GUAACAAUCACUUGUGACGCAUUUAACCCAUUGUCUCCCGC-UGGACGGAGGCGUUCAUGUGGGCAAUUAACACCUUCUCCU---UCGGAUGCGAAAUGCCCC---CUUUUGUUACAUUU ((((((((((....)))(((((.......((((...(((((-((((((....)))))).)))))))))............(.---...)))))).........---...))))))).... ( -29.20) >DroSec_CAF1 431 119 + 1 GUAACAAUCACCAGUGACGCAUUUAACCCAAUGUCUCCCGC-UGGACGGAGGCGUUCAUGUGGGCAAUUAACACCUUCUCCUUCUUCGGAUGCGAAAUGCCCCCUCCUUUUGUUACAUUU (((((((...((((((..(((((......)))))....)))-)))..(((((.........(((((.((..((.....(((......)))))..)).))))))))))..))))))).... ( -34.70) >DroSim_CAF1 2627 119 + 1 GUAACAAUCACCAGUGACGCAUUUAACCCAUUGUCUCCCGC-UGGACGGAGGCGUUCAUGUGGGCAAUUAACACCUUCUCCUUCUUCAGAUGCGAAAUGCCCCCUCCUUUUGUUACAUUU (((((((...((((((..(((..........)))....)))-)))..(((((.........(((((.((..((...(((........)))))..)).))))))))))..))))))).... ( -30.70) >DroEre_CAF1 395 113 + 1 GUAACAAUCAGUCGUGACGCAUUUAACCCAUUGUCUCCCGCCUGGACGGAGGCGUUCAUGUGGGCAAUUAACACCUUCUCCU---UCAGAUGCGAAAUGCC-C---CUUUUGUUACAUUU (((((((...(.(((..(((((((.((.....))..(((((.((((((....)))))).)))))..................---..)))))))..))).)-.---...))))))).... ( -29.20) >DroYak_CAF1 427 114 + 1 GUAACAAUCAGUUGUGACGCAUUUAACCCAUUGUCUCCCGCCUGGACGGAGGCGUUCAUGUGGGCAAUUAACACCUUCUCCU---GCAGAUGCGAAAUGCCCC---CUUUUGUUACAUUU ((((((((((....))).((((((.....((((...(((((.((((((....)))))).))))))))).............(---((....)))))))))...---...))))))).... ( -28.80) >consensus GUAACAAUCACUAGUGACGCAUUUAACCCAUUGUCUCCCGC_UGGACGGAGGCGUUCAUGUGGGCAAUUAACACCUUCUCCU___UCAGAUGCGAAAUGCCCC___CUUUUGUUACAUUU ((((((((((....)))(((((((.((.....))..(((((.((((((....)))))).))))).......................)))))))...............))))))).... (-28.28 = -28.04 + -0.24)

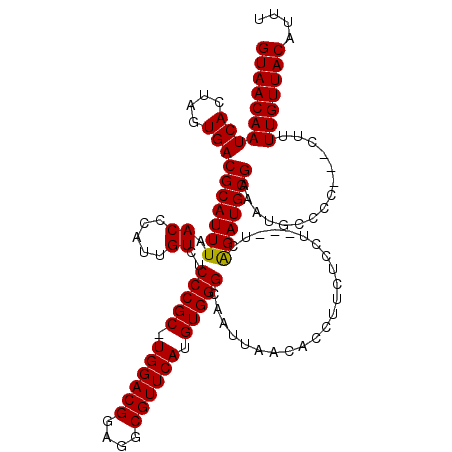
| Location | 15,891,914 – 15,892,027 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.21 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -28.96 |
| Energy contribution | -28.92 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15891914 113 - 23771897 AAAUGUAACAAAAG---GGGGCAUUUCGCAUCCGA---AGGAGAAGGUGUUAAUUGCCCACAUGAACGCCUCCGUCCA-GCGGGAGACAAUGGGUUAAAUGCGUCACAAGUGAUUGUUAC ....(((((((...---.(((((.((.((((((..---..)....))))).)).))))).(((..((((.....((((-...(....)..))))......)))).....))).))))))) ( -31.00) >DroSec_CAF1 431 119 - 1 AAAUGUAACAAAAGGAGGGGGCAUUUCGCAUCCGAAGAAGGAGAAGGUGUUAAUUGCCCACAUGAACGCCUCCGUCCA-GCGGGAGACAUUGGGUUAAAUGCGUCACUGGUGAUUGUUAC ....(((((((..((((((((((...(((.(((......)))....))).....))))).(......))))))...((-.(((((..((((......))))..)).))).)).))))))) ( -37.10) >DroSim_CAF1 2627 119 - 1 AAAUGUAACAAAAGGAGGGGGCAUUUCGCAUCUGAAGAAGGAGAAGGUGUUAAUUGCCCACAUGAACGCCUCCGUCCA-GCGGGAGACAAUGGGUUAAAUGCGUCACUGGUGAUUGUUAC ....(((((((..((((((((((.((.((((((...........)))))).)).))))).(......))))))...((-.(((..(((.((.......))..))).))).)).))))))) ( -35.70) >DroEre_CAF1 395 113 - 1 AAAUGUAACAAAAG---G-GGCAUUUCGCAUCUGA---AGGAGAAGGUGUUAAUUGCCCACAUGAACGCCUCCGUCCAGGCGGGAGACAAUGGGUUAAAUGCGUCACGACUGAUUGUUAC ....(((((((.((---(-((((.((.((((((..---......)))))).)).))))).(.(((.(((.(((((....))))).(((.....)))....)))))).).))..))))))) ( -35.00) >DroYak_CAF1 427 114 - 1 AAAUGUAACAAAAG---GGGGCAUUUCGCAUCUGC---AGGAGAAGGUGUUAAUUGCCCACAUGAACGCCUCCGUCCAGGCGGGAGACAAUGGGUUAAAUGCGUCACAACUGAUUGUUAC ....(((((((..(---((((((.((.((((((..---......)))))).)).)))))...(((.(((.(((((....))))).(((.....)))....))))))...))..))))))) ( -35.60) >consensus AAAUGUAACAAAAG___GGGGCAUUUCGCAUCUGA___AGGAGAAGGUGUUAAUUGCCCACAUGAACGCCUCCGUCCA_GCGGGAGACAAUGGGUUAAAUGCGUCACAAGUGAUUGUUAC ....(((((((.......(((((...(((.(((......)))....))).....)))))...(((.(((.(((((....))))).(((.....)))....)))))).......))))))) (-28.96 = -28.92 + -0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:43 2006