
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,882,249 – 15,882,351 |
| Length | 102 |
| Max. P | 0.701667 |

| Location | 15,882,249 – 15,882,351 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.53 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15882249 102 - 23771897 UUU-U-----------------UCUGGCUUGCUGCUUUUGCAUUUUGCGUGUUGAUCUAAGCGGUUUAUUAGCCCCGAUUUGCAAGUAAGCUGCUUAACAUCACCCCCACCACGCUGGUC ...-.-----------------.(..((..(((((....)))....))(((.((((.(((((((((((((.((........)).)))))))))))))..))))....)))...))..).. ( -27.40) >DroPse_CAF1 12061 116 - 1 GGCCAGCUGACAGUUGACUGCUGCCUGCUGCCUGCUUUUGCAUUUUGCGUGUUGAUCUAAGCGGUUUAUUAGCCCCGAUUUGCAAGUAAGCUGCUUAGCAGGGGC----CCAUUCUGGCC ((((((.((.((((.....))))...(((.(((((....((.....)).........(((((((((((((.((........)).)))))))))))))))))))))----.))..)))))) ( -42.30) >DroSec_CAF1 27398 103 - 1 UUUCA-----------------UCUGGCUUGCUGCUUUUGCAUUUUGCGUGUUGAUCUAAGCGGUUUAUUAGCCCCGAUUUGCAAGUAAGCUGCUUAACAUCACCCCCACCAUGCUGGUC .....-----------------...((((.(((((....)))...((.(((.((((.(((((((((((((.((........)).)))))))))))))..))))....))))).)).)))) ( -27.20) >DroEre_CAF1 20397 101 - 1 UUU-------------------UCUGGGUUCUUGCUUUUGCAUUUUGCGUGUUGAUCUAAGCGGUUUAUUAGCCACGAUUUGCGAGUAAGCUGCUUAGCAUCUCCGCUUCCACGCUGGUC ...-------------------..(((((...(((....)))....))(((..(((((((((((((((((.((........)).)))))))))))))).)))..)))..)))........ ( -31.20) >DroYak_CAF1 30449 101 - 1 UUU-------------------UCUGGUUUGCUUCUUUUGCAUUUUGCGUGUUGAUCUAAGCGGUUUAUUAGCCAAGCUUUGCAAGUAAGUAGCUUUACAUCGGCCCCUCCACACUGGAU ...-------------------((..((.(((((..((((((....((..(((......)))((((....))))..))..)))))).)))))(((.......)))........))..)). ( -21.20) >DroAna_CAF1 11070 98 - 1 ----------------------UCUGGUUUGCUGCUUUUGCAUUUUGCGUGUUGAUCUAAGCGGUUUAUUAGCCCCGAUUUACAAGUAAGCCGCUUAACAUUGCCCGCACCAGGCUGCCC ----------------------((((((....(((....)))....(((.((.....(((((((((((((..............))))))))))))).....)).)))))))))...... ( -29.24) >consensus UUU___________________UCUGGCUUGCUGCUUUUGCAUUUUGCGUGUUGAUCUAAGCGGUUUAUUAGCCCCGAUUUGCAAGUAAGCUGCUUAACAUCGCCCCCACCACGCUGGUC .......................(((((...........((.....))((((.....(((((((((((((.((........)).)))))))))))))))))............))))).. (-17.00 = -17.53 + 0.53)

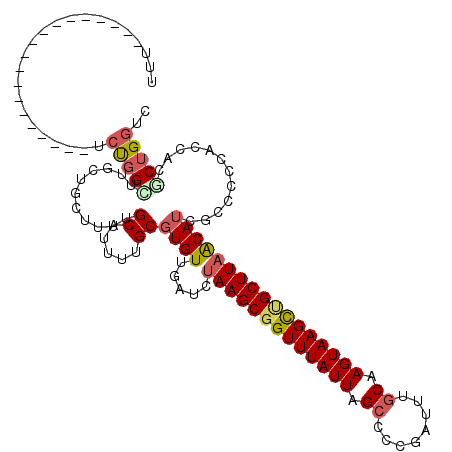
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:37 2006