
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,880,857 – 15,880,966 |
| Length | 109 |
| Max. P | 0.884423 |

| Location | 15,880,857 – 15,880,966 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 94.19 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -26.48 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
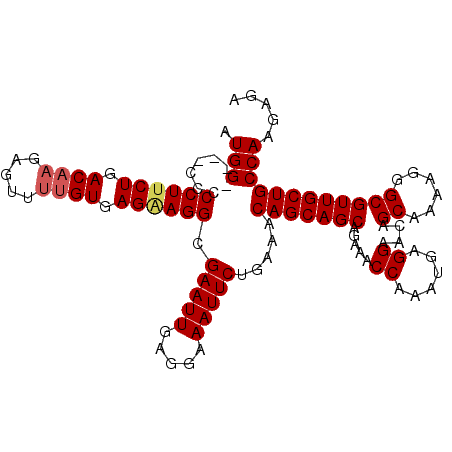
>3L_DroMel_CAF1 15880857 109 + 23771897 AUGG-GCUCCCCCUUCUGACAAGAGUUUUGUGAGAAGGCGAAUUGAGGAAAUUCUGAAACAGCAGCAGAAACCAAAUGAGGAACAGCAAAAGGGCGUUGCUGCCAAGAGC ....-((((..((((((.(((((...))))).)))))).(((((.....))))).....(((((((.....((......))....((......)))))))))....)))) ( -31.40) >DroSec_CAF1 25944 110 + 1 AUGGGGCACCCCCUUCUGACAAGAGUUUUGUGAGAAGGGGAAUUGAGGAAAUUCUGAAACAGCAGCAGAAACCAAAUGAGGAACAGCAAAAGGGCGUUGCUGCCAAGAGC ....(((..((((((((.(((((...))))).))))))))....................((((((.....((......))....((......)))))))))))...... ( -35.20) >DroEre_CAF1 18963 106 + 1 AUGG----CCCCCUCCUGACAAGAGUUCUGUGAGAAGGCGAAUUGAGGAAAUUCUGAAACAGCAGCAGAAACCAAAUGAGGAACAGCAAAAGGGCGUUGCUGCCAAGGGA ....----..((((.((.(((.(....)))).))..((((((((.....)))))......((((((.....((......))....((......))))))))))).)))). ( -27.60) >DroYak_CAF1 28942 106 + 1 AUGG----CCCCCUUCUGACAAGAGUUUCGUGAGGAGGCGAAUUGAGGAAAUUCUGAAACAGCAGCAGAAACCAAAUGAGGAACAGCAAAAGGGCGUUGCUGCCAAGAGA ...(----((.((((.(((........))).)))).)))(((((.....))))).....(((((((.....((......))....((......)))))))))........ ( -27.50) >consensus AUGG____CCCCCUUCUGACAAGAGUUUUGUGAGAAGGCGAAUUGAGGAAAUUCUGAAACAGCAGCAGAAACCAAAUGAGGAACAGCAAAAGGGCGUUGCUGCCAAGAGA .(((.......((((((.((((.....)))).)))))).(((((.....))))).....(((((((.....((......))....((......))))))))))))..... (-26.48 = -27.10 + 0.62)

| Location | 15,880,857 – 15,880,966 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 94.19 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -24.96 |
| Energy contribution | -24.52 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15880857 109 - 23771897 GCUCUUGGCAGCAACGCCCUUUUGCUGUUCCUCAUUUGGUUUCUGCUGCUGUUUCAGAAUUUCCUCAAUUCGCCUUCUCACAAAACUCUUGUCAGAAGGGGGAGC-CCAU (((((((((((((..((......))....((......))....)))))))......(((((.....))))).((((((.((((.....)))).))))))))))))-.... ( -31.60) >DroSec_CAF1 25944 110 - 1 GCUCUUGGCAGCAACGCCCUUUUGCUGUUCCUCAUUUGGUUUCUGCUGCUGUUUCAGAAUUUCCUCAAUUCCCCUUCUCACAAAACUCUUGUCAGAAGGGGGUGCCCCAU ((....(((((((..((......))....((......))....)))))))...................(((((((((.((((.....)))).))))))))).))..... ( -30.70) >DroEre_CAF1 18963 106 - 1 UCCCUUGGCAGCAACGCCCUUUUGCUGUUCCUCAUUUGGUUUCUGCUGCUGUUUCAGAAUUUCCUCAAUUCGCCUUCUCACAGAACUCUUGUCAGGAGGGGG----CCAU .(((..(((((((..((......))....((......))....)))))))......(((((.....))))).((((((.((((.....)))).)))))))))----.... ( -27.20) >DroYak_CAF1 28942 106 - 1 UCUCUUGGCAGCAACGCCCUUUUGCUGUUCCUCAUUUGGUUUCUGCUGCUGUUUCAGAAUUUCCUCAAUUCGCCUCCUCACGAAACUCUUGUCAGAAGGGGG----CCAU ......(((((((..((......))....((......))....)))))))......(((((.....)))))((((((((((((.....))))..).))))))----)... ( -25.90) >consensus GCUCUUGGCAGCAACGCCCUUUUGCUGUUCCUCAUUUGGUUUCUGCUGCUGUUUCAGAAUUUCCUCAAUUCGCCUUCUCACAAAACUCUUGUCAGAAGGGGG____CCAU .(((..(((((((..((......))....((......))....)))))))......(((((.....))))).((((((.((((.....)))).)))))))))........ (-24.96 = -24.52 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:36 2006