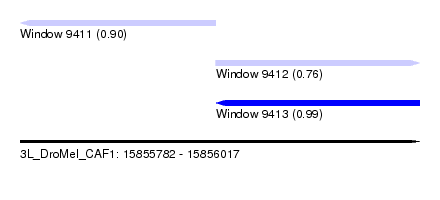
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,855,782 – 15,856,017 |
| Length | 235 |
| Max. P | 0.993322 |

| Location | 15,855,782 – 15,855,897 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -24.74 |
| Energy contribution | -25.17 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15855782 115 - 23771897 CGGAGCAAUGCUGGCCACGUCAUGAAUGACUGCCCAUCGAAUGCUCAAAUGCUCGAUUGCCAAGCGGAAAAAGUGCUGAGAAAAGCGGUAGCUCAAAAAACUGAAAAGGAAAA-AA ..((((..(((((((...((((....)))).....(((((..((......))))))).))).)))).....(.((((......)))).).))))......((....)).....-.. ( -28.40) >DroSim_CAF1 64610 116 - 1 CGGAGCAAUGCUGGCCACGUCAUGAAUGACUGCCCAUCGAAUGCUCAAAUGCUCGAAUGCCAAGCGGAAAAAGUGCUGAGAAAAGCGGUAGCUCAAAAAACUGAAAAGAAAAAAAA ..((((..(((((((...((((....))))......((((..((......))))))..))).)))).....(.((((......)))).).))))......((....))........ ( -27.80) >DroEre_CAF1 62728 88 - 1 CGGAGCAAUGCUGGCCACGUCAUGAAUGACUGCCCAUCGAGUG--------CUCGAGUGCCAAGCGGAAAAAUUGCUGAGAAAAGCGGUAGCUCAA-------------------- ..((((..(((((((.((((((....))))......((((...--------.))))))))).)))).....((((((......)))))).))))..-------------------- ( -28.80) >DroYak_CAF1 104357 103 - 1 CGGAGCAAUGCUGGCCACGUCAUGAAUGACUGCCCAUCGAAGG--------CUCGAAUGCCAAGCGGAAAAACUGCUGAGAAAAGCGGUAGCUCAAACAACUGAAAAGGAA----- ..((((..((((((.((.((((....)))))).))......((--------(......))).)))).....((((((......)))))).))))......((....))...----- ( -30.80) >consensus CGGAGCAAUGCUGGCCACGUCAUGAAUGACUGCCCAUCGAAUG________CUCGAAUGCCAAGCGGAAAAAGUGCUGAGAAAAGCGGUAGCUCAAAAAACUGAAAAGGAA_____ ..((((..(((((((...((((....))))......((((............))))..))).)))).....((((((......)))))).))))...................... (-24.74 = -25.17 + 0.44)


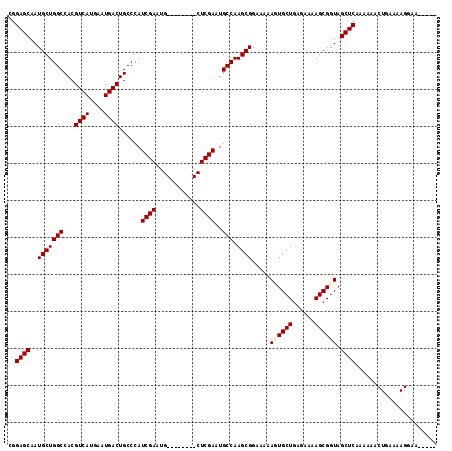
| Location | 15,855,897 – 15,856,017 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -28.54 |
| Energy contribution | -28.65 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15855897 120 + 23771897 GCCUAAUCCUCCACUGCCGUUUUGCCUGCCAAAUCGGCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGCGCUAUUUGGCAGGCUUUUGCCUGCUUUUCAUGGUUUAC ...........(((.((((.((((.....)))).)))).)))((((.(((.................(((((((.....)))))))...(((((((....)))))))))).))))..... ( -36.70) >DroPse_CAF1 61801 120 + 1 GCGUAAUCCUCGACUGCCGCUUUGCCUGCCAAAUCGUCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGUGCUAUUUGGCAGGCUUUUGUCUGCGCUUCAAGGCUUAC ..((((.(((.((..((.((...(((((((((((.......((((......................))))((((.....)))).)))))))))))........)))).)).))).)))) ( -37.35) >DroSim_CAF1 64726 120 + 1 GCCUAAUCCUCUACUGCCGUUUUGCCUGCCAAAUCGGCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGCGCUAUUUGGCAGGCUUUUGUCUGCUUUUCAUGGUUUAC ...(((.((......((.(....((((((((((..(((...))).......................(((((((.....)))))))))))))))))......).)).......)).))). ( -33.22) >DroEre_CAF1 62816 120 + 1 GCCUAAUCCUCUACUGCCGUUUUGCCUGCCAAAUCGGCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGCGCUAUUUGGCAGGCUUUUGUCUGCUUUUCAGCGUUUAC ...............((((.((((.....)))).))))(((((((......................)))))))......(((((....(((((((....)))))))....))))).... ( -34.25) >DroYak_CAF1 104460 120 + 1 GCCUAAUCCUCCACUGCCGUUUUGCCUGCCAAAUCGGCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGCGCUAUUUGGCAGGCUUUUGUCUGCUGUUCAGGGUUUAC ....((((((..((.((.(....((((((((((..(((...))).......................(((((((.....)))))))))))))))))......).)).))..))))))... ( -38.50) >DroPer_CAF1 63692 120 + 1 GCGUAAUCCUCGACUGCCGCUUUGCCUGCCAAAUCGUCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGUGCUAUUUGGCAGGCUUUUGUCUGCGCUUCAAGGCUUAC ..((((.(((.((..((.((...(((((((((((.......((((......................))))((((.....)))).)))))))))))........)))).)).))).)))) ( -37.35) >consensus GCCUAAUCCUCCACUGCCGUUUUGCCUGCCAAAUCGGCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGCGCUAUUUGGCAGGCUUUUGUCUGCUUUUCAAGGUUUAC ...............((.(....((((((((((..(((...))).......................(((((((.....)))))))))))))))))......).)).............. (-28.54 = -28.65 + 0.11)

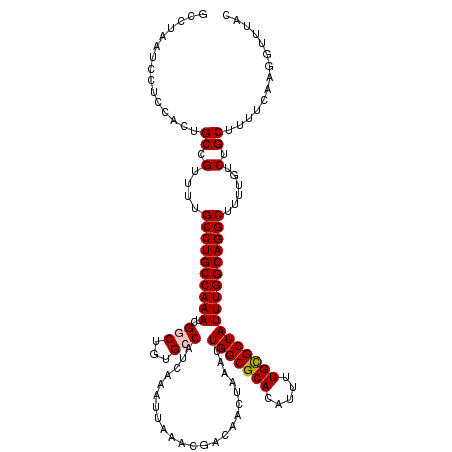

| Location | 15,855,897 – 15,856,017 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -40.17 |
| Consensus MFE | -34.03 |
| Energy contribution | -33.81 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15855897 120 - 23771897 GUAAACCAUGAAAAGCAGGCAAAAGCCUGCCAAAUAGCGCAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGCCGAUUUGGCAGGCAAAACGGCAGUGGAGGAUUAGGC .....((((.....((((((....))))(((.....((((.....))))(((((.(..(((((((((......)))))))..))..).))))).)))......)).)))).......... ( -37.30) >DroPse_CAF1 61801 120 - 1 GUAAGCCUUGAAGCGCAGACAAAAGCCUGCCAAAUAGCACAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGACGAUUUGGCAGGCAAAGCGGCAGUCGAGGAUUACGC ((((.((((((...((........(((((((((((.((((.....))))((((.((.(((((.....))))).)))))).......)))))))))))......))..)))))).)))).. ( -45.74) >DroSim_CAF1 64726 120 - 1 GUAAACCAUGAAAAGCAGACAAAAGCCUGCCAAAUAGCGCAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGCCGAUUUGGCAGGCAAAACGGCAGUAGAGGAUUAGGC .....((.................(((((((((((.(((((.....))))).......((..(((.......)))..)).......)))))))))))...((....))...))....... ( -34.90) >DroEre_CAF1 62816 120 - 1 GUAAACGCUGAAAAGCAGACAAAAGCCUGCCAAAUAGCGCAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGCCGAUUUGGCAGGCAAAACGGCAGUAGAGGAUUAGGC ((....(((....)))........(((((((((((.(((((.....))))).......((..(((.......)))..)).......)))))))))))......))............... ( -38.40) >DroYak_CAF1 104460 120 - 1 GUAAACCCUGAACAGCAGACAAAAGCCUGCCAAAUAGCGCAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGCCGAUUUGGCAGGCAAAACGGCAGUGGAGGAUUAGGC ......(((..((.((........(((((((((((.(((((.....))))).......((..(((.......)))..)).......)))))))))))......)).))..)))....... ( -38.94) >DroPer_CAF1 63692 120 - 1 GUAAGCCUUGAAGCGCAGACAAAAGCCUGCCAAAUAGCACAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGACGAUUUGGCAGGCAAAGCGGCAGUCGAGGAUUACGC ((((.((((((...((........(((((((((((.((((.....))))((((.((.(((((.....))))).)))))).......)))))))))))......))..)))))).)))).. ( -45.74) >consensus GUAAACCAUGAAAAGCAGACAAAAGCCUGCCAAAUAGCGCAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGCCGAUUUGGCAGGCAAAACGGCAGUAGAGGAUUAGGC ..............((........(((((((((((.((((.....))))((((.((.(((((.....))))).)))))).......)))))))))))......))............... (-34.03 = -33.81 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:26 2006